| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:40:04 UTC |
|---|
| Update Date | 2020-05-21 16:28:59 UTC |
|---|
| BMDB ID | BMDB0001024 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Phosphohydroxypyruvic acid |
|---|
| Description | Phosphohydroxypyruvic acid, also known as 3-phosphooxypyruvate or hydroxypyruvic acid phosphate, belongs to the class of organic compounds known as glycerone phosphates. These are organic compounds containing a glycerone moiety that carries a phosphate group at the O-1 or O-2 position. Phosphohydroxypyruvic acid is a moderately acidic compound (based on its pKa). Phosphohydroxypyruvic acid exists in all living species, ranging from bacteria to humans. |
|---|
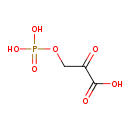
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Phosphohydroxypyruvic acid | ChEBI | | Hydroxypyruvic acid phosphate | ChEBI | | 3-Phosphonooxypyruvic acid | Kegg | | 3-Phosphohydroxypyruvate | Kegg | | 3-Phosphooxypyruvate | Kegg | | Hydroxypyruvate phosphate | Generator | | Hydroxypyruvic acid phosphoric acid | Generator | | 3-Phosphonooxypyruvate | Generator | | 3-Phosphooxypyruvic acid | Generator | | Phosphohydroxypyruvate | Generator | | 2-oxo-3-(Phosphonooxy)-propanoate | HMDB | | 2-oxo-3-(Phosphonooxy)-propanoic acid | HMDB | | 3-Phosphonatooxypyruvate | HMDB | | 2-Oxo-3-(phosphonooxy)propanoic acid | HMDB | | 2-Oxo-3-(phosphonooxy)propionic acid | HMDB | | Phosphohydroxypyruvic acid | HMDB |
|
|---|
| Chemical Formula | C3H5O7P |
|---|
| Average Molecular Weight | 184.0414 |
|---|
| Monoisotopic Molecular Weight | 183.977289026 |
|---|
| IUPAC Name | 2-oxo-3-(phosphonooxy)propanoic acid |
|---|
| Traditional Name | phosphohydroxypyruvate |
|---|
| CAS Registry Number | 3913-50-6 |
|---|
| SMILES | OC(=O)C(=O)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H5O7P/c4-2(3(5)6)1-10-11(7,8)9/h1H2,(H,5,6)(H2,7,8,9) |
|---|
| InChI Key | LFLUCDOSQPJJBE-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycerone phosphates. These are organic compounds containing a glycerone moiety that carries a phosphate group at the O-1 or O-2 position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Glycerone phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycerone phosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Monosaccharide
- Keto acid
- Alpha-keto acid
- Alpha-hydroxy ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9300000000-33ff06735f8214248ea3 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00xr-9510000000-f67b2feb11cbb37fdf47 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-4411ba8f6713e6b68f94 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00s9-0900000000-457227604cff5cdecb73 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05g0-6900000000-5ced177e050ba3272f0f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003r-3900000000-22820af5ec79da81207b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9300000000-0127ef53c9426c592779 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4ba77bd112070310c576 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-1900000000-0d44a3e3fb04adf7390b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000f-9400000000-29a2afefebab0a9dc88b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0016-9000000000-2f054e3a4d1b04f903f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9500000000-d989c4c4b7671c11809a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-7e6d85bfb63dfb4e3ea0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|