| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:40:22 UTC |
|---|
| Update Date | 2020-05-21 16:28:25 UTC |
|---|
| BMDB ID | BMDB0001049 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Gamma-Glutamylcysteine |
|---|
| Description | gamma-Glutamylcysteine, also known as L-γ-glutamylcysteine or gamma-glu-cys, belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. gamma-Glutamylcysteine is a very strong basic compound (based on its pKa). gamma-Glutamylcysteine exists in all living species, ranging from bacteria to humans. gamma-Glutamylcysteine is a potentially toxic compound. |
|---|
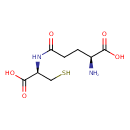
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-L-Glutamyl-L-cysteine | ChEBI | | gamma-Glu-cys | ChEBI | | gamma-L-Glutamyl-L-cysteine | ChEBI | | L-gamma-Glutamylcysteine | ChEBI | | g-Glu-cys | Generator | | Γ-glu-cys | Generator | | g-L-Glutamyl-L-cysteine | Generator | | Γ-L-glutamyl-L-cysteine | Generator | | L-g-Glutamylcysteine | Generator | | L-Γ-glutamylcysteine | Generator | | g-Glutamylcysteine | Generator | | Γ-glutamylcysteine | Generator | | Γ-L-glu-L-cys | HMDB | | L-Γ-glutamyl-L-cysteine | HMDB | | N-Γ-glutamylcysteine | HMDB | | N-L-Γ-glutamylcysteine | HMDB | | N-L-Γ-glutamyl-L-cysteine | HMDB | | gamma-L-Glu-L-cys | HMDB | | L-gamma-Glutamyl-L-cysteine | HMDB | | N-gamma-Glutamylcysteine | HMDB | | N-L-gamma-Glutamylcysteine | HMDB | | N-L-gamma-Glutamyl-L-cysteine | HMDB | | 5-L-Glutamylcysteine | HMDB | | H-gamma-Glu-cys-OH | HMDB | | L-g-Glutamyl-L-cysteine | HMDB | | N-Γ-L-glutamyl-L-cysteine | HMDB | | gamma-Glutamylcysteine | HMDB |
|
|---|
| Chemical Formula | C8H14N2O5S |
|---|
| Average Molecular Weight | 250.272 |
|---|
| Monoisotopic Molecular Weight | 250.062342258 |
|---|
| IUPAC Name | (2S)-2-amino-4-{[(1R)-1-carboxy-2-sulfanylethyl]carbamoyl}butanoic acid |
|---|
| Traditional Name | gamma-glutamylcysteine |
|---|
| CAS Registry Number | 636-58-8 |
|---|
| SMILES | N[C@@H](CCC(=O)N[C@@H](CS)C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C8H14N2O5S/c9-4(7(12)13)1-2-6(11)10-5(3-16)8(14)15/h4-5,16H,1-3,9H2,(H,10,11)(H,12,13)(H,14,15)/t4-,5-/m0/s1 |
|---|
| InChI Key | RITKHVBHSGLULN-WHFBIAKZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-acyl-alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-alpha-amino acid
- Cysteine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Fatty acid
- Dicarboxylic acid or derivatives
- Amino acid
- Alkylthiol
- Carboximidic acid
- Carboximidic acid derivative
- Carboxylic acid
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Organosulfur compound
- Primary aliphatic amine
- Primary amine
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Amine
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-054t-1920100000-5ce9a082971db6d9e2e9 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-054t-1920100000-5ce9a082971db6d9e2e9 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-053u-9630000000-344be03dbe2132b00c88 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00b9-8494000000-88b1eea1e1badaee084b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udi-0090000000-50bd1555ededc1afde80 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0udi-0290000000-385ca433f03af7530f47 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0ue9-5490000000-95d45a9cd2bb6249d3a2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9510000000-5f29ca903abe05256c83 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-001i-9510000000-5f29ca903abe05256c83 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0002-1490000000-bfce673369358b8079e9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0690000000-c49357f0d28683125b6d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-002b-2490000000-41c54aabf6acd4d3b53e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-014j-9050000000-1137fb3c62232376669c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fr-1790000000-2aaa1344429a2297cc6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-4920000000-4ad42780853d5599c05b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05fr-9400000000-b67fd0eaf646aa8f47b3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000t-1390000000-e723899e4ca71bda7600 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00sj-3960000000-9a2f96ad7d24e7546315 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00e9-9300000000-e14b428bbb5b63d3ae06 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00b9-0910000000-302a05e4e453b27bad64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-3900000000-da869ba294aa1d1e7c49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-9100000000-4bc7ff3637ea175068f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0zmi-0960000000-675f718188f5caad73ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9600000000-13ddc5ab8de093dd11ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9100000000-8b8a5550fa1112649784 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|