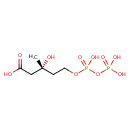| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:40:58 UTC |
|---|
| Update Date | 2020-05-21 16:27:03 UTC |
|---|
| BMDB ID | BMDB0001090 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5-Diphosphomevalonic acid |
|---|
| Description | (R)-5-Diphosphomevalonic acid, also known as mevalonate 5-diphosphate or mevalonic acid 5-pyrophosphate, belongs to the class of organic compounds known as organic pyrophosphates. These are organic compounds containing the pyrophosphate oxoanion, with the structure OP([O-])(=O)OP(O)([O-])=O (R)-5-Diphosphomevalonic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral (R)-5-Diphosphomevalonic acid exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (R)-5-Diphosphomevalonate | ChEBI | | 5-Diphosphomevalonate | HMDB | | 5-Diphosphomevalonic acid | HMDB | | Mevalonate 5-diphosphate | HMDB | | Mevalonate pyrophosphate | HMDB | | Mevalonate-diphosphate | HMDB | | (3R)-3-Hydroxy-5-[[hydroxy(phosphonooxy)phosphinyl]oxy]-3-methylpentanoic acid | HMDB | | (R)-Diphosphomevalonic acid | HMDB | | 5-Pyrophosphomevalonic acid | HMDB | | Mevalonic 5-pyrophosphate | HMDB | | Mevalonic acid 5-diphosphate | HMDB | | Mevalonic acid 5-pyrophosphate | HMDB | | Mevalonic acid pyrophosphate | HMDB | | Pyrophosphomevalonic acid | HMDB | | (3R)-3-Hydroxy-5-[[hydroxy(phosphonooxy)phosphinyl]oxy]-3-methylpentanoate | HMDB | | (R)-Diphosphomevalonate | HMDB | | 5-Pyrophosphomevalonate | HMDB | | Mevalonate 5-pyrophosphate | HMDB | | Mevalonic 5-pyrophosphic acid | HMDB | | Mevalonic acid 5-diphosphic acid | HMDB | | Mevalonic acid 5-pyrophosphic acid | HMDB | | Mevalonic acid pyrophosphic acid | HMDB | | Pyrophosphomevalonate | HMDB | | (R)-5-Diphosphomevalonic acid | Generator | | R-Diphosphomevalonate | HMDB |
|
|---|
| Chemical Formula | C6H14O10P2 |
|---|
| Average Molecular Weight | 308.1169 |
|---|
| Monoisotopic Molecular Weight | 308.006219692 |
|---|
| IUPAC Name | (3R)-3-hydroxy-5-{[hydroxy(phosphonooxy)phosphoryl]oxy}-3-methylpentanoic acid |
|---|
| Traditional Name | mevalonate-diphosphate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | C[C@@](O)(CCOP(O)(=O)OP(O)(O)=O)CC(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H14O10P2/c1-6(9,4-5(7)8)2-3-15-18(13,14)16-17(10,11)12/h9H,2-4H2,1H3,(H,7,8)(H,13,14)(H2,10,11,12)/t6-/m1/s1 |
|---|
| InChI Key | SIGQQUBJQXSAMW-ZCFIWIBFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as organic pyrophosphates. These are organic compounds containing the pyrophosphate oxoanion, with the structure OP([O-])(=O)OP(O)([O-])=O. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organic oxoanionic compounds |
|---|
| Sub Class | Organic pyrophosphates |
|---|
| Direct Parent | Organic pyrophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Organic pyrophosphate
- Monoalkyl phosphate
- Short-chain hydroxy acid
- Organic phosphoric acid derivative
- Fatty acid
- Alkyl phosphate
- Phosphoric acid ester
- Tertiary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Carbonyl group
- Alcohol
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004m-9820000000-7d8dd5ca92f4eb0538fd | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00dv-9243200000-6d8177d69adb810da37f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01wf-1391000000-a8414ad19cb44452295f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03e9-4940000000-c856f917d05bbc77ac8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01sr-9840000000-db467c852b0f7d795ece | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0bt9-1494000000-7e72bc5c055d7b3e4f37 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-7920000000-5c36505c154e9a3abebd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-cbcfedc91b4fe71d5849 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0019000000-a7e7073b139ac0acc855 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a6r-9466000000-5ab0d4ae0def99221d8f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056s-9500000000-38a7f595bafdd824dd36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a59-0938000000-9afa28886c9f8ff753e0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-3940000000-53f4c41d4cec2fbfc65d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08n9-6910000000-e20806c8945d9a51ddb1 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|