| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:41:55 UTC |
|---|
| Update Date | 2020-05-21 16:28:59 UTC |
|---|
| BMDB ID | BMDB0001167 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Pyruvaldehyde |
|---|
| Description | Pyruvaldehyde, also known as 2-oxopropanal or 1,2-propanedione, belongs to the class of organic compounds known as alpha ketoaldehydes. These are organic compounds containing an aldehyde substituted with a keto group on the adjacent carbon. Pyruvaldehyde exists in all living species, ranging from bacteria to plants to humans. Based on a literature review a significant number of articles have been published on Pyruvaldehyde. |
|---|
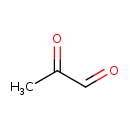
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2-Propanedione | ChEBI | | 2-Ketopropionaldehyde | ChEBI | | 2-Oxopropanal | ChEBI | | 2-Oxopropionaldehyde | ChEBI | | Acetylformaldehyde | ChEBI | | Acetylformyl | ChEBI | | alpha-Ketopropionaldehyde | ChEBI | | CH3COCHO | ChEBI | | Pyruvic aldehyde | ChEBI | | a-Ketopropionaldehyde | Generator | | Α-ketopropionaldehyde | Generator | | Aldehyde, pyruvic | MeSH | | Oxopropanal | MeSH | | 1-Ketopropionaldehyde | HMDB | | 2-keto Propionaldehyde | HMDB | | 2-oxo-Propionaldehyde | HMDB | | Ketopropionaldehyde | HMDB | | Methylglyoxal | HMDB | | Propanedione | HMDB | | Propanolone | HMDB | | Pyroracemic aldehyde | HMDB |
|
|---|
| Chemical Formula | C3H4O2 |
|---|
| Average Molecular Weight | 72.0627 |
|---|
| Monoisotopic Molecular Weight | 72.021129372 |
|---|
| IUPAC Name | 2-oxopropanal |
|---|
| Traditional Name | methylglyoxal |
|---|
| CAS Registry Number | 78-98-8 |
|---|
| SMILES | CC(=O)C=O |
|---|
| InChI Identifier | InChI=1S/C3H4O2/c1-3(5)2-4/h2H,1H3 |
|---|
| InChI Key | AIJULSRZWUXGPQ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha ketoaldehydes. These are organic compounds containing an aldehyde substituted with a keto group on the adjacent carbon. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Alpha ketoaldehydes |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-ketoaldehyde
- Ketone
- Organic oxide
- Hydrocarbon derivative
- Short-chain aldehyde
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | < 25 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9000000000-43f9b3c94058c64733b5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00dj-9000000000-964129275940a60a617d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-006y-9000000000-0ae1e5fd2d50b28f967c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00dj-9000000000-964129275940a60a617d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-9000000000-110d6fcd891f2c54a2cb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05fr-9000000000-bec3651f9ea6825cf4f7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-8db5b1ba128748e220c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-9000000000-f968a2358e6fd85ae268 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9000000000-0797bdeaa575b54943c5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uk9-9000000000-89ef1c082b2ef672eb67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-9000000000-5b936b9153956c2f64be | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9000000000-9190b139940adf0d92c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-570ca33f905cecb028a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9000000000-482389dce4f05675f174 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-79267be0bc618dc8018f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-87bbaed151efac084591 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|