| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:16 UTC |
|---|
| Update Date | 2020-05-21 16:29:05 UTC |
|---|
| BMDB ID | BMDB0001187 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 1D-Myo-inositol 1,3,4,6-tetrakisphosphate |
|---|
| Description | 1D-Myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate, also known as 1d-myo-inositol 1,3,4,6-tetrakisphosphate, belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. 1D-Myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate is possibly soluble (in water) and an extremely strong acidic compound (based on its pKa). 1D-Myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate exists in all eukaryotes, ranging from yeast to humans. 1D-Myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate participates in a number of enzymatic reactions, within cattle. In particular, 1D-Myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate can be biosynthesized from inositol 1,3,4-trisphosphate; which is catalyzed by the enzyme inositol-tetrakisphosphate 1-kinase. In addition, 1D-Myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate can be converted into 1D-myo-inositol 3,4-bisphosphate through the action of the enzyme inositol polyphosphate 1-phosphatase. In cattle, 111d-myo-1d-myo-inositol 1,3,4,6-tetrakisphosphate is involved in the metabolic pathway called the inositol metabolism pathway. |
|---|
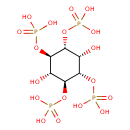
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1D-Myo-inositol 1,3,4,6-tetrakisphosphate | ChEBI | | Inositol 1,3,4,6-tetrakisphosphate | ChEBI | | Inositol-1,3,4,6-tetrakisphosphate | ChEBI | | Inositol-1,3,4,6-tetraphosphate | ChEBI | | Ins-1,3,4,6-P4 | ChEBI | | Myo-inositol-1,3,4,6-tetrakisphosphate | ChEBI | | 1D-Myo-inositol 1,3,4,6-tetrakisphosphoric acid | Generator | | Inositol 1,3,4,6-tetrakisphosphoric acid | Generator | | Inositol-1,3,4,6-tetrakisphosphoric acid | Generator | | Inositol-1,3,4,6-tetraphosphoric acid | Generator | | Myo-inositol-1,3,4,6-tetrakisphosphoric acid | Generator | | D-Myo-inositol 1,3,4,6-tetrakisphosphoric acid | Generator | | D-Myo-inositol 1,3,4,5-tetrakis-*phosphate potass | HMDB | | D-Myo-inositol-1,3,4,5-tetrakisphosphate octapotassium salt | HMDB | | Inositol 1,3,4,6-tetrakis(phosphate) | HMDB | | Inositol 1,3,4,6-tetraphosphate | HMDB | | Ins(1,3,4,6)P4 | HMDB | | D-Myo-inositol 1,3,4,6-tetraphosphate | HMDB | | D-myo-Inositol 1,3,4,6-tetrakisphosphate | ChEBI | | D-myo-Inositol (1,3,4,6)-tetrakisphosphate | HMDB |
|
|---|
| Chemical Formula | C6H16O18P4 |
|---|
| Average Molecular Weight | 500.0755 |
|---|
| Monoisotopic Molecular Weight | 499.928709756 |
|---|
| IUPAC Name | {[(1R,2s,3S,4S,5r,6R)-2,5-dihydroxy-3,4,6-tris(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | [(1R,2s,3S,4S,5r,6R)-2,5-dihydroxy-3,4,6-tris(phosphonooxy)cyclohexyl]oxyphosphonic acid |
|---|
| CAS Registry Number | 110298-84-5 |
|---|
| SMILES | O[C@H]1[C@H](OP(O)(O)=O)[C@@H](OP(O)(O)=O)[C@@H](O)[C@@H](OP(O)(O)=O)[C@@H]1OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H16O18P4/c7-1-3(21-25(9,10)11)5(23-27(15,16)17)2(8)6(24-28(18,19)20)4(1)22-26(12,13)14/h1-8H,(H2,9,10,11)(H2,12,13,14)(H2,15,16,17)(H2,18,19,20)/t1-,2+,3-,4+,5-,6+ |
|---|
| InChI Key | ZAWIXNGTTZTBKV-JMVOWJSSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Monoalkyl phosphate
- Cyclohexanol
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9130200000-72a8edec18faf8e95f8e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-006t-8714394000-de66a4c41082c3293325 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-3000490000-928166c23ec83db57e33 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ue9-3001890000-c68d476bcc2e35c523ba | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-6129000000-c6bd3f736cfac8d67b21 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-4000900000-4a9f3e003f338f491753 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000300000-46dfa53774ee48631c2d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-d6c1739743ae930182a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000190000-0b94f644b1d50a849c1c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000490000-d8ea5bdd4162c42be27d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9100000000-08da9faffb9fd54b245e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000900000-43744c672ffce8e5f032 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0032-3000900000-57364afe1c2a5bb935e9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-6f311a985bc6bd348fa8 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|