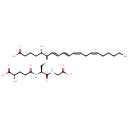| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:28 UTC |
|---|
| Update Date | 2020-05-19 22:01:11 UTC |
|---|
| BMDB ID | BMDB0001198 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Leukotriene C4 |
|---|
| Description | Leukotriene C4, also known as 5S,6R-LTC(sub 4) or LTC4, belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. Thus, leukotriene C4 is considered to be an eicosanoid lipid molecule. Leukotriene C4 is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Within humans, leukotriene C4 participates in a number of enzymatic reactions. In particular, leukotriene C4 can be biosynthesized from leukotriene A4 and glutathione; which is catalyzed by the enzyme leukotriene C4 synthase. In addition, leukotriene C4 can be converted into leukotriene D4 and L-glutamic acid through its interaction with the enzyme Gamma-glutamyltranspeptidase 1. In humans, leukotriene C4 is involved in mefenamic acid action pathway. Outside of the human body, Leukotriene C4 has been detected, but not quantified in, several different foods, such as romaine lettuces, mung beans, annual wild rices, dandelions, and java plums. This could make leukotriene C4 a potential biomarker for the consumption of these foods. Leukotriene C4, with regard to humans, has been found to be associated with several diseases such as leukotriene c4-synthesis deficiency, aseptic meningitis, hydrocephalus, and aids; leukotriene C4 has also been linked to the inborn metabolic disorder glutathione synthetase deficiency. A leukotriene that is (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid in which a glutathionyl group is attached at position 6 via a sulfide linkage. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (R-(R*,s*-(e,e,Z,Z)))-N-(S-(1-(4-carboxy-1-hydroxybutyl)-2,4,6,9-pentadecatetraenyl)-N-L-gamma-glutamyl-L-cysteinyl)glycine | ChEBI | | 5S,6R-LTC(Sub 4) | ChEBI | | 5S-Hydroxy,6R-(S-glutathionyl),7E,9E,11Z,14Z-eicosatetraenoic acid | ChEBI | | LTC (Sub 4) | ChEBI | | LTC4 | ChEBI | | (R-(R*,s*-(e,e,Z,Z)))-N-(S-(1-(4-carboxy-1-hydroxybutyl)-2,4,6,9-pentadecatetraenyl)-N-L-g-glutamyl-L-cysteinyl)glycine | Generator | | (R-(R*,s*-(e,e,Z,Z)))-N-(S-(1-(4-carboxy-1-hydroxybutyl)-2,4,6,9-pentadecatetraenyl)-N-L-γ-glutamyl-L-cysteinyl)glycine | Generator | | 5S-Hydroxy,6R-(S-glutathionyl),7E,9E,11Z,14Z-eicosatetraenoate | Generator | | Leukotriene C-1 | HMDB | | Leukotriene C-4 | HMDB | | Leukotrienes C | HMDB | | Leukotriene C | HMDB | | Leukotriene C 1 | HMDB | | Leukotriene C 4 | HMDB | | Leukotriene C1 | HMDB | | (R-(R*,s*-(e,e,Z,Z)))-N-(S-(1-(4-carboxy-1-hydroxybutyl)-2,4,6,9-pentadecatetraenyl)-N-L-gamma-glutamyl-L-cysteinyl)-glycine | HMDB | | 5S,6R-LTC | HMDB | | L-gamma-Glutamyl-S-[(1R,2E,4E,6Z,9Z)-1-[(1S)-4-carboxy-1-hydroxybutyl]-2,4,6,9-pentadecatetraenyl]-L-cysteinyl-glycine | HMDB | | Leucotriene C4 | HMDB | | LTC | HMDB | | [R-[R*,s*-(e,e,Z,Z)]]-N-[S-[1-(4-carboxy-1-hydroxybutyl)-2,4,6,9-pentadecatetraenyl]-N-L-gamma-glutamyl-L-cysteinyl]-glycine 5S,6R-LTC4 | HMDB | | 5S,6R-LTC4 | HMDB | | Leukotriene C4 | HMDB |
|
|---|
| Chemical Formula | C30H47N3O9S |
|---|
| Average Molecular Weight | 625.774 |
|---|
| Monoisotopic Molecular Weight | 625.303300807 |
|---|
| IUPAC Name | (5S,6R,7E,9E,11Z,14Z)-6-{[(2R)-2-[(4S)-4-amino-4-carboxybutanamido]-2-[(carboxymethyl)carbamoyl]ethyl]sulfanyl}-5-hydroxyicosa-7,9,11,14-tetraenoic acid |
|---|
| Traditional Name | leukotriene C4 |
|---|
| CAS Registry Number | 72025-60-6 |
|---|
| SMILES | CCCCC\C=C/C\C=C/C=C/C=C/[C@@H](SC[C@H](NC(=O)CC[C@H](N)C(O)=O)C(=O)NCC(O)=O)[C@@H](O)CCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C30H47N3O9S/c1-2-3-4-5-6-7-8-9-10-11-12-13-16-25(24(34)15-14-17-27(36)37)43-21-23(29(40)32-20-28(38)39)33-26(35)19-18-22(31)30(41)42/h6-7,9-13,16,22-25,34H,2-5,8,14-15,17-21,31H2,1H3,(H,32,40)(H,33,35)(H,36,37)(H,38,39)(H,41,42)/b7-6-,10-9-,12-11+,16-13+/t22-,23-,24-,25+/m0/s1 |
|---|
| InChI Key | GWNVDXQDILPJIG-NXOLIXFESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Oligopeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-oligopeptide
- Gamma-glutamyl alpha peptide
- Leukotriene
- Hydroxyeicosatetraenoic acid
- Eicosanoid
- Glutamine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Cysteine or derivatives
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- L-alpha-amino acid
- Alpha-amino acid
- N-substituted-alpha-amino acid
- Tricarboxylic acid or derivatives
- Hydroxy fatty acid
- Thia fatty acid
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Secondary alcohol
- Amino acid
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Dialkylthioether
- Carboxylic acid
- Sulfenyl compound
- Thioether
- Primary amine
- Organic oxygen compound
- Organic nitrogen compound
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Amine
- Organic oxide
- Organopnictogen compound
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05o0-3202091000-c6dee8507ab8b61b0c02 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0g5c-6203049000-51753829f5101ccfd684 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_17) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-00di-0000009000-9ba4d29fb234bf629c19 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-00di-0000009000-c2e3b5fb18c749bec683 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-00di-0000009000-764950659a3ede43e5fa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-00di-0000009000-84d0f40152dc35593991 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-00di-0000009000-2b9de1e9017bddef56ff | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-00di-0020019000-c37ade6bbbbb0db63082 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-0c00-0290044000-556a3c7aef20c3fd1ba2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-0un9-0492001000-2801dff9e15f1a96421c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QIT , negative | splash10-01t9-0970000000-e6b8d4063abe84fe54f6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0c34-2010394000-b2537b2e21446a86fab1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9303360000-546ae5580086593ee435 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9013010000-43aff9255ff94b18ed35 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0kn9-0127079000-3855e84af2520307dddf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zgr-0139010000-fdb78d5f775d532f53ec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f6y-2912000000-a86e557888d76eea9df8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0010009000-78d18a02a7849f6bf4ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-1951014000-2f6a645de70656f26d29 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0007-6900010000-a4f76d24eb6f5ec03a33 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0570-1405119000-3fe7d1ddf1fa60edcdb7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-1902000000-cf43af95f58742e02019 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05o0-2900000000-92e9d6fced329468ccfd | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|