| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:41 UTC |
|---|
| Update Date | 2020-04-22 15:07:10 UTC |
|---|
| BMDB ID | BMDB0001216 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
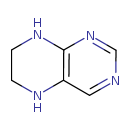
| Common Name | Tetrahydropteridine |
|---|
| Description | Tetrahydropteridine, also known as H4-biopterin or THP, belongs to the class of organic compounds known as pteridines and derivatives. These are polycyclic aromatic compounds containing a pteridine moiety, which consists of a pyrimidine fused to a pyrazine ring to form pyrimido(4,5-b)pyrazine. Tetrahydropteridine is a very strong basic compound (based on its pKa). Tetrahydropteridine exists in all living organisms, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5,6,7,8-Tetrahydropteridine | Kegg | | 5,6,7,8-Tetrahydro-pteridine | HMDB | | H4-Biopterin | HMDB | | THP | HMDB | | Tetrahydropteridine | ChEBI |
|
|---|
| Chemical Formula | C6H8N4 |
|---|
| Average Molecular Weight | 136.1545 |
|---|
| Monoisotopic Molecular Weight | 136.074896276 |
|---|
| IUPAC Name | 5,6,7,8-tetrahydropteridine |
|---|
| Traditional Name | 5,6,7,8-tetrahydropteridine |
|---|
| CAS Registry Number | 10593-78-9 |
|---|
| SMILES | C1CNC2=C(N1)C=NC=N2 |
|---|
| InChI Identifier | InChI=1S/C6H8N4/c1-2-9-6-5(8-1)3-7-4-10-6/h3-4,8H,1-2H2,(H,7,9,10) |
|---|
| InChI Key | IDAICLIJTRXNER-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pteridines and derivatives. These are polycyclic aromatic compounds containing a pteridine moiety, which consists of a pyrimidine fused to a pyrazine ring to form pyrimido(4,5-b)pyrazine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Pteridines and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pteridine
- Secondary aliphatic/aromatic amine
- Imidolactam
- Pyrimidine
- Heteroaromatic compound
- Azacycle
- Secondary amine
- Organic nitrogen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organonitrogen compound
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-2900000000-f10ca50f06250e5030d3 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0900000000-9a1ed9296c96ac152018 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-63f9acde2a431bff2488 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-016r-9100000000-9987b7952a5c983f7c8e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0900000000-93507d1f946deac8a074 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0900000000-36867a49c6c36342b032 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-9800000000-c2d528a7546845512953 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0900000000-278b0289a2d1c7916e5b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0900000000-7f4a76469d170f8d43f9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05nf-7900000000-e829877fe8774c9c13f9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0900000000-06cd9080747456c7643d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-c5f7233447b8e475f5c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9500000000-a22ac8b5a57e8c2437f7 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Taylor, E. C., Jr.; Carbon, John A.; Garland, Robert B.; Hoff, Dale R.; Howell, Charles F.; Sherman, Wm. R. Recent developments in pteridine synthesis. Ciba Foundation Symposium, Chem. and Biol. Pteridines (1954), 104-20. |
|---|