| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:48 UTC |
|---|
| Update Date | 2020-05-20 23:10:19 UTC |
|---|
| BMDB ID | BMDB0001229 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Dopaquinone |
|---|
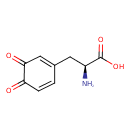
| Description | Dopaquinone belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. Dopaquinone is possibly soluble (in water) and a very strong basic compound (based on its pKa). Dopaquinone exists in all living organisms, ranging from bacteria to humans. Dopaquinone can be biosynthesized from L-dopa through the action of the enzyme tyrosinase. In cattle, dopaquinone is involved in the metabolic pathway called the tyrosine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2-Amino-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)propanoate | ChEBI | | (S)-2-Amino-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)propanoic acid | Generator | | O-Dopaquinone | HMDB | | 4-(2-Carboxy-2-aminoethyl)-1,2-benzoquinone | HMDB | | Dopaquinone, (S)-isomer | HMDB | | L-Dopaquinone | HMDB |
|
|---|
| Chemical Formula | C9H9NO4 |
|---|
| Average Molecular Weight | 195.1721 |
|---|
| Monoisotopic Molecular Weight | 195.053157781 |
|---|
| IUPAC Name | (2S)-2-amino-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)propanoic acid |
|---|
| Traditional Name | dopaquinone |
|---|
| CAS Registry Number | 4430-97-1 |
|---|
| SMILES | N[C@@H](CC1=CC(=O)C(=O)C=C1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C9H9NO4/c10-6(9(13)14)3-5-1-2-7(11)8(12)4-5/h1-2,4,6H,3,10H2,(H,13,14)/t6-/m0/s1 |
|---|
| InChI Key | AHMIDUVKSGCHAU-LURJTMIESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- O-benzoquinone
- Quinone
- Ketone
- Amino acid
- Cyclic ketone
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Amine
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fml-5900000000-3f77b48d5f42d8951324 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9570000000-5d835cfbd2b202044cb7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ufs-0900000000-093c511b443d87916930 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ue9-1900000000-fc3bdef4314080b351c1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uxr-9200000000-07d0c6318c984ef80e41 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-7481c7c6fe6294da272c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002f-0900000000-04347396e1171463fb62 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-7900000000-13ce3640966908fffe55 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-c319fd057861dc7d7ad4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00e9-3900000000-b64033d6c2ec2ca4314b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-5900000000-cde5f7d52f36428ed06e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udj-0900000000-2ca1383c9487e79be90a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ff0-0900000000-92f4bf729acf4be5b6bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00e9-8900000000-d3972df871a20993fe05 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|