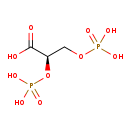| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:42 UTC |
|---|
| Update Date | 2020-05-19 22:01:13 UTC |
|---|
| BMDB ID | BMDB0001294 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 2,3-Diphosphoglyceric acid |
|---|
| Description | 2,3-Diphosphoglyceric acid, also known as 2,3-disphospho-D-glycerate or DPG, belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. 2,3-Diphosphoglyceric acid is an extremely strong acidic compound (based on its pKa). 2,3-Diphosphoglyceric acid exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,3-Bisphospho-D-glycerate | ChEBI | | 2,3-Bisphosphoglyceric acid | ChEBI | | 2,3-BPG | ChEBI | | 2,3-Disphospho-D-glycerate | ChEBI | | D-Greenwald ester | ChEBI | | DPG | ChEBI | | Glyceric acid bis(dihydrogen phosphate) | ChEBI | | Glyceric acid diphosphate | ChEBI | | 2,3-Bisphospho-D-glyceric acid | Generator | | 2,3-Bisphosphoglycerate | Generator | | 2,3-Disphospho-D-glyceric acid | Generator | | Glycerate bis(dihydrogen phosphate) | Generator | | Glyceric acid bis(dihydrogen phosphoric acid) | Generator | | Glycerate diphosphate | Generator | | Glyceric acid diphosphoric acid | Generator | | 2,3-Diphosphoglycerate | Generator | | (2R)-2,3-Bis(phosphonooxy)-propanoate | HMDB | | (2R)-2,3-Bis(phosphonooxy)-propanoic acid | HMDB | | (R)-2,3-Bis(phosphonooxy)-propanoate | HMDB | | (R)-2,3-Bis(phosphonooxy)-propanoic acid | HMDB | | 2,3-Bis(phosphonooxy)-propanoate | HMDB | | 2,3-Bis(phosphonooxy)-propanoic acid | HMDB | | 2,3-Diphospho-D-glycerate | HMDB | | 2,3-Diphospho-D-glyceric acid | HMDB | | 2,3-Diphospho-D-glyceric acid pentasodium salt | HMDB | | D-Glyceric acid bis | HMDB | | D-Glyceric acid bis(dihydrogen phosphate) | HMDB | | Diphosphoglycerate | HMDB | | Diphosphoglyceric acid | HMDB | | Glycerate 2,3-diphosphate | HMDB | | 2,3 Diphosphoglyceric acid | HMDB | | 2,3 Bisphosphoglycerate | HMDB | | 2,3 Diphosphoglycerate | HMDB | | 2,3-DPG | HMDB | | 2,3-Diphosphoglycerate, (D)-isomer | HMDB | | Glycerate 2,3-bisphosphate | HMDB | | 2,3-Bisphosphate, glycerate | HMDB | | (2R)-2,3-Bis(phosphonooxy)propanoic acid | HMDB | | (2R)-2,3-Bis(phosphonooxy)propionic acid | HMDB | | 2,3-Bis(phosphonooxy)propanoic acid | HMDB | | 2,3-Bis(phosphonooxy)propionic acid | HMDB | | 2,3-Diphosphoglyceric acid | HMDB | | 2,3-Diphosphonooxypropanoic acid | HMDB | | 2,3-Diphosphonooxypropionic acid | HMDB |
|
|---|
| Chemical Formula | C3H8O10P2 |
|---|
| Average Molecular Weight | 266.0371 |
|---|
| Monoisotopic Molecular Weight | 265.9592695 |
|---|
| IUPAC Name | (2R)-2,3-bis(phosphonooxy)propanoic acid |
|---|
| Traditional Name | diphosphoglycerate |
|---|
| CAS Registry Number | 138-81-8 |
|---|
| SMILES | OC(=O)[C@@H](COP(O)(O)=O)OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H8O10P2/c4-3(5)2(13-15(9,10)11)1-12-14(6,7)8/h2H,1H2,(H,4,5)(H2,6,7,8)(H2,9,10,11)/t2-/m1/s1 |
|---|
| InChI Key | XOHUEYCVLUUEJJ-UWTATZPHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Sugar acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monoalkyl phosphate
- Glyceric_acid
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Monosaccharide
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9320000000-fbd8c78fd88d592fab56 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-006t-9410000000-ea4c253779009c08e1bd | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-014i-0930000000-5127f7f7568a961a8078 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00di-2900000000-e9e3c8a32ece4b7f9fce | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-006t-9300000000-e09861cf36df495519f6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-014i-0090000000-4a9ded352d5912810741 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-066r-1960000000-0d244402e55e5ef9de1d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0ar0-5960000000-1d5a4352115c6c5eb82f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-056r-9330000000-70ae76c58e470fe3b594 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-004i-9100000000-6cacbc2756ef980dc5a2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0uxr-1490000000-ddf502cfcfd4ff0bca0b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-00xr-6910000000-871a376b75658c914a1e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00xr-9200000000-ab1b55ce1ad971f1b3f8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-016r-9100000000-2af5a77daca9f7cef852 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-014i-9100000000-1d6a9786ab78002b7ac2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-014i-0090000000-4a9ded352d5912810741 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-066r-1960000000-fcab8021959f67dc5a44 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0ar0-5960000000-1d5a4352115c6c5eb82f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-056r-9330000000-70ae76c58e470fe3b594 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9100000000-37f228e9721638e33212 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0uxr-1490000000-ddf502cfcfd4ff0bca0b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-1930000000-6b2743652b0e2aa81960 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00r2-8890000000-64b460dbcc7be6d3e820 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-6900000000-7cbb7d86341bbdc1500a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03fr-5090000000-73e5bca24796a45bebaf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9010000000-a07defbdfd47a3f23225 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-9f9abb79cd423f7b15f0 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|