| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:43 UTC |
|---|
| Update Date | 2020-05-21 16:28:37 UTC |
|---|
| BMDB ID | BMDB0001295 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
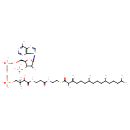
| Common Name | 2-Hydroxyphytanoyl-CoA |
|---|
| Description | 2-Hydroxyphytanoyl-CoA, also known as 3S2hphy-CoA, belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. Thus, 2-hydroxyphytanoyl-CoA is considered to be a fatty ester lipid molecule. 2-Hydroxyphytanoyl-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3S)-2-Hydroxyphytanoyl-CoA | HMDB | | (3S)-2-Hydroxyphytanoyl-coenzyme A | HMDB | | (3S,7R,11R)-2-Hydroxy-3,7,11,15-tetramethylhexadecanoyl-CoA | HMDB | | (3S,7R,11R)-2-Hydroxy-3,7,11,15-tetramethylhexadecanoyl-coenzyme A | HMDB | | 2-Hydroxyphytanoyl-coenzyme A | HMDB | | 3(S)-2-Hydroxyphytanoyl-CoA | HMDB | | 3(S)-2-Hydroxyphytanoyl-coenzyme A | HMDB | | 3S2hPhy-CoA | HMDB | | 3S2hPhy-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C41H74N7O18P3S |
|---|
| Average Molecular Weight | 1078.049 |
|---|
| Monoisotopic Molecular Weight | 1077.402388825 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({hydroxy[3-hydroxy-3-({2-[(2-{[(3S,7R,11R)-2-hydroxy-3,7,11,15-tetramethylhexadecanoyl]sulfanyl}ethyl)carbamoyl]ethyl}carbamoyl)-2,2-dimethylpropoxy]phosphoryl}oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-[({hydroxy[hydroxy(3-hydroxy-3-({2-[(2-{[(3S,7R,11R)-2-hydroxy-3,7,11,15-tetramethylhexadecanoyl]sulfanyl}ethyl)carbamoyl]ethyl}carbamoyl)-2,2-dimethylpropoxy)phosphoryl]oxyphosphoryl}oxy)methyl]oxolan-3-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | 172787-73-4 |
|---|
| SMILES | CC(C)CCC[C@@H](C)CCC[C@@H](C)CCC[C@H](C)C(O)C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C41H74N7O18P3S/c1-25(2)11-8-12-26(3)13-9-14-27(4)15-10-16-28(5)32(50)40(54)70-20-19-43-30(49)17-18-44-38(53)35(52)41(6,7)22-63-69(60,61)66-68(58,59)62-21-29-34(65-67(55,56)57)33(51)39(64-29)48-24-47-31-36(42)45-23-46-37(31)48/h23-29,32-35,39,50-52H,8-22H2,1-7H3,(H,43,49)(H,44,53)(H,58,59)(H,60,61)(H2,42,45,46)(H2,55,56,57)/t26-,27-,28+,29-,32?,33-,34-,35?,39-/m1/s1 |
|---|
| InChI Key | WNVFJMYPVBOLKV-PJDIVXIPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose monosaccharide
- Tetrahydrofuran
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Ether
- Dialkyl ether
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
- Mitochondria
- Peroxisome
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-4901110200-170aee27b4e79c002131 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-1913130000-f3f3a31b1f2f1eadfa1b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1900010000-8676dc6b110ef7b1cd90 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05rr-8732251900-bda6cb0d556520fc587c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0560-5932220200-8cfcacf29ba06547d671 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a7i-5900100000-5c05acebab4cee45d967 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-9000000101-0d2b8860e57967864001 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-059i-9200000014-1db94d0080dd1e75278c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-0110290000-a0e067a6ba49b57c31dd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9010000100-2c73fb7c13f83d7ae4f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9002101200-89e778b8abfee3306a19 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056r-9002402700-78e6d232a30f300def33 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|