| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:53 UTC |
|---|
| Update Date | 2020-05-21 16:27:07 UTC |
|---|
| BMDB ID | BMDB0001308 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5'-Phosphoribosyl-N-formylglycinamide |
|---|
| Description | 5'-Phosphoribosyl-N-formylglycinamide, also known as N-formylglycinamide ribonucleotide or N-formyl-gar, belongs to the class of organic compounds known as glycinamide ribonucleotides. Glycinamide ribonucleotides are compounds in which the amide N atom of glycineamide is linked to the C-1 of a ribosyl (or deoxyribosyl) moiety. Nucleotides have a phosphate group linked to the C5 carbon of the ribose (or deoxyribose) moiety. 5'-Phosphoribosyl-N-formylglycinamide is an extremely weak basic (essentially neutral) compound (based on its pKa). 5'-Phosphoribosyl-N-formylglycinamide exists in all living species, ranging from bacteria to humans. |
|---|
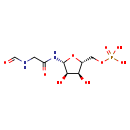
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N-Formyl-gar | Kegg | | N-Formylglycinamide ribonucleotide | Kegg | | N2-Formyl-N1-(5-phospho-D-ribosyl)glycinamide | Kegg | | 5'-p-Ribosyl-N-formylglycineamide | HMDB | | 5'-Phosphoribosyl-formylglycinamide | HMDB | | 5'-Phosphoribosyl-N-formylglycineamide | HMDB | | 5-Phosphoribosyl-N-formalglycineamide | HMDB | | FGAR | HMDB | | N(2)-Formyl-N(1)-(5-phospho-D-ribosyl)glycinamide | HMDB | | Formylglycinamide ribonucleotide | HMDB | | Formylglycineamideribotide | HMDB | | Phosphoribosyl-N-formylglycineamide | HMDB | | 2-(Formylamino)-N-(5-O-phosphono-beta-D-ribofuranosyl)acetamide | HMDB | | 2-(Formylamino)-N-(5-O-phosphono-β-D-ribofuranosyl)acetamide | HMDB | | 5'-Phosphoribosyl-N-formylglycinamide | HMDB | | 5’-Phosphoribosyl-N-formylglycinamide | HMDB | | Formylglycinamide ribotide | HMDB | | Phosphoribosylformylglycinamide | HMDB | | alpha-N-Formylglycinamide ribonucleotide | HMDB | | α-N-Formylglycinamide ribonucleotide | HMDB |
|
|---|
| Chemical Formula | C8H15N2O9P |
|---|
| Average Molecular Weight | 314.1865 |
|---|
| Monoisotopic Molecular Weight | 314.0515166 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-3,4-dihydroxy-5-(2-formamidoacetamido)oxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-3,4-dihydroxy-5-(2-formamidoacetamido)oxolan-2-yl]methoxyphosphonic acid |
|---|
| CAS Registry Number | 349-34-8 |
|---|
| SMILES | O[C@H]1[C@@H](O)[C@H](NC(=O)CNC=O)O[C@@H]1COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C8H15N2O9P/c11-3-9-1-5(12)10-8-7(14)6(13)4(19-8)2-18-20(15,16)17/h3-4,6-8,13-14H,1-2H2,(H,9,11)(H,10,12)(H2,15,16,17)/t4-,6-,7-,8-/m1/s1 |
|---|
| InChI Key | VDXLUNDMVKSKHO-XVFCMESISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycinamide ribonucleotides. Glycinamide ribonucleotides are compounds in which the amide N atom of glycineamide is linked to the C-1 of a ribosyl (or deoxyribosyl) moiety. Nucleotides have a phosphate group linked to the C5 carbon of the ribose (or deoxyribose) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Glycinamide ribonucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Glycinamide ribonucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycinamide-ribonucleotide
- Pentose phosphate
- Pentose-5-phosphate
- N-formyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- N-formyl-alpha-amino acid
- N-substituted-alpha-amino acid
- Alpha-amino acid or derivatives
- Monosaccharide phosphate
- Pentose monosaccharide
- Monoalkyl phosphate
- Organic phosphoric acid derivative
- Alkyl phosphate
- Monosaccharide
- Phosphoric acid ester
- Tetrahydrofuran
- Secondary carboxylic acid amide
- 1,2-diol
- Carboxamide group
- Secondary alcohol
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Alcohol
- Organic nitrogen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9330000000-47bd3a51454a22349c16 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0089-5972200000-28cbe677e79c994d5692 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-9121000000-70e7f7e392d67c9062d7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9110000000-24b14d3360fd37fa0b5a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9100000000-6c2f243f9e36f7267013 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01r6-8914000000-411b5cf47e3e7e5511fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9200000000-fc9464d8ad7b3b1ac820 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-ccc93e5e04249a7f1425 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0079000000-0c457b0a6a5c25c65962 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-2920000000-569fd550eacc61a8bd6c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-2900000000-32e3418d15835aba1b7b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01t9-9036000000-b8480e9ce9f70df638e9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-14698f4417bb19153972 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-e93ac01407977ed80740 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|