| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:45:58 UTC |
|---|
| Update Date | 2020-05-19 22:01:26 UTC |
|---|
| BMDB ID | BMDB0001439 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Phosphoribosyl formamidocarboxamide |
|---|
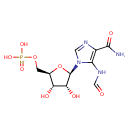
| Description | Phosphoribosyl formamidocarboxamide, also known as FAICAR, belongs to the class of organic compounds known as 1-ribosyl-imidazolecarboxamides. These are organic compounds containing the imidazole ring linked to a ribose ring through a 1-2 bond. Phosphoribosyl formamidocarboxamide exists in all living species, ranging from bacteria to plants to humans. Based on a literature review very few articles have been published on Phosphoribosyl formamidocarboxamide. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-(5'-Phosphoribosyl)-5-formamido-4-imidazolecarboxamide | ChEBI | | 5'-Phosphoribosyl-5-formamido-4-imidazolecarboxamide | ChEBI | | 5-Formamido-1-(5-phosphoribosyl)imidazole-4-carboxamide | ChEBI | | FAICAR | ChEBI | | 5-Formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide | Kegg | | 5'-P-Ribosyl-5-formamido-4-imidazole carboxamide | HMDB | | 5'-Phosphoribosyl-5-formamido-4-imidazole carboxamide | HMDB | | 5-amino-1-(5-phospho-D-Ribosyl)imidazole-4-carboxamide | HMDB | | 5-formamido-1-(5-Phosphoribosyl)-imidazole-4-carboxamide | HMDB | | 5-Phosphoribosyl-5-formamido-4-imid-carboxamide | HMDB | | Phosphoribosyl-formamido-carboxamide | HMDB | | 5-Formamidoimidazole-4-carboxamide ribotide | MeSH, HMDB | | 5-(Formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide | HMDB | | 5-(Formylamino)-1-(5-O-phosphono-β-D-ribofuranosyl)-1H-imidazole-4-carboxamide | HMDB | | 5-Formylamino-4-imidazolecarboxamide ribonucleotide | HMDB |
|
|---|
| Chemical Formula | C10H15N4O9P |
|---|
| Average Molecular Weight | 366.2213 |
|---|
| Monoisotopic Molecular Weight | 366.05766461 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(4-carbamoyl-5-formamido-1H-imidazol-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(4-carbamoyl-5-formamidoimidazol-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxyphosphonic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC(=O)C1=C(NC=O)N(C=N1)[C@@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C10H15N4O9P/c11-8(18)5-9(13-3-15)14(2-12-5)10-7(17)6(16)4(23-10)1-22-24(19,20)21/h2-4,6-7,10,16-17H,1H2,(H2,11,18)(H,13,15)(H2,19,20,21)/t4-,6-,7-,10-/m1/s1 |
|---|
| InChI Key | ABCOOORLYAOBOZ-KQYNXXCUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-ribosyl-imidazolecarboxamides. These are organic compounds containing the imidazole ring linked to a ribose ring through a 1-2 bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Imidazole ribonucleosides and ribonucleotides |
|---|
| Sub Class | 1-ribosyl-imidazolecarboxamides |
|---|
| Direct Parent | 1-ribosyl-imidazolecarboxamides |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-ribosyl-imidazolecarboxamide
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Pentose monosaccharide
- N-arylamide
- 2-heteroaryl carboxamide
- Imidazole-4-carbonyl group
- Monoalkyl phosphate
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Alkyl phosphate
- Phosphoric acid ester
- Heteroaromatic compound
- Azole
- Vinylogous amide
- Imidazole
- Tetrahydrofuran
- Carboxamide group
- 1,2-diol
- Secondary carboxylic acid amide
- Primary carboxylic acid amide
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxide
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Organic oxygen compound
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9725000000-466b9ed7af43597f7fff | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0002-9240300000-b9e749599e4fdfe0510a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ar0-0914000000-1d61e2aa1204766a006e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-0900000000-051245189eb5cd0ff26f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-1900000000-f18e08c70bdaa9aa6913 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0gdj-8809000000-e71e875c5ef95845f25d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fb9-9700000000-1930562d45d841bd928d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9100000000-5b37e0fb5e34ee3423f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-016u-9007000000-60a18b9e82325eba8d6a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9002000000-45c1e86e798b498a86e9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-681d7ff850cbe4448fe9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0819000000-600f59ce04e9f2036927 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000j-1922000000-ab410f3897dc2f486680 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bt9-4900000000-cd0a95ff69a87468beeb | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|