| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:47:26 UTC |
|---|
| Update Date | 2020-05-11 20:53:28 UTC |
|---|
| BMDB ID | BMDB0001542 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N-Acetyllactosamine |
|---|
| Description | N-Acetyllactosamine, also known as galb1-4glcnacb or lacnac, belongs to the class of organic compounds known as acylaminosugars. These are organic compounds containing a sugar linked to a chain through N-acyl group. N-Acetyllactosamine is an extremely weak basic (essentially neutral) compound (based on its pKa). N-Acetyllactosamine exists in all living organisms, ranging from bacteria to humans. |
|---|
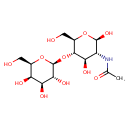
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| beta-D-Galactosyl-1,4-N-acetyl-D-glucosamine | ChEBI | | Galb1-4glcnacb | ChEBI | | Galbeta1-4glcnacbeta | ChEBI | | LacNAc | ChEBI | | N-Acetyl-beta-lactosamine | ChEBI | | beta-D-Galactosyl-1,4-N-acetyl-beta-D-glucosamine | Kegg | | b-D-Galactosyl-1,4-N-acetyl-D-glucosamine | Generator | | Β-D-galactosyl-1,4-N-acetyl-D-glucosamine | Generator | | N-Acetyl-b-lactosamine | Generator | | N-Acetyl-β-lactosamine | Generator | | b-D-Galactosyl-1,4-N-acetyl-b-D-glucosamine | Generator | | Β-D-galactosyl-1,4-N-acetyl-β-D-glucosamine | Generator | | beta-delta-Galactosyl-1,4-N-acetyl-beta-delta-glucosamine | HMDB | | beta-delta-Galactosyl-1,4-N-acetyl-delta-glucosamine | HMDB | | D-Galactopyranosyl | HMDB | | delta-Galactopyranosyl | HMDB | | N-Acetyl-lactosamine | HMDB | | ACETYL lactosamine | HMDB |
|
|---|
| Chemical Formula | C14H25NO11 |
|---|
| Average Molecular Weight | 383.3484 |
|---|
| Monoisotopic Molecular Weight | 383.142760647 |
|---|
| IUPAC Name | N-[(2R,3R,4R,5S,6R)-2,4-dihydroxy-6-(hydroxymethyl)-5-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-3-yl]acetamide |
|---|
| Traditional Name | N-acetyllactosamine |
|---|
| CAS Registry Number | 32181-59-2 |
|---|
| SMILES | CC(=O)N[C@H]1[C@H](O)O[C@H](CO)[C@@H](O[C@@H]2O[C@H](CO)[C@H](O)[C@H](O)[C@H]2O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C14H25NO11/c1-4(18)15-7-9(20)12(6(3-17)24-13(7)23)26-14-11(22)10(21)8(19)5(2-16)25-14/h5-14,16-17,19-23H,2-3H2,1H3,(H,15,18)/t5-,6-,7-,8+,9-,10+,11-,12-,13-,14+/m1/s1 |
|---|
| InChI Key | KFEUJDWYNGMDBV-LODBTCKLSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acylaminosugars. These are organic compounds containing a sugar linked to a chain through N-acyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Acylaminosugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acylaminosugar
- N-acyl-alpha-hexosamine
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Oxane
- Acetamide
- Carboxamide group
- Hemiacetal
- Secondary carboxylic acid amide
- Secondary alcohol
- Acetal
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Organic nitrogen compound
- Organopnictogen compound
- Alcohol
- Primary alcohol
- Organonitrogen compound
- Carbonyl group
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | - beta-D-galactopyranosyl-(1->4)-N-acetyl-D-glucosamine (CHEBI:16153 )
|
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0uy3-5539000000-9001b4ffdf2686f61fdb | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0a4i-3410139000-490a44b54a788af499ad | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0g4i-0294000000-eba3eabd1634e1f899d6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-002r-0900000000-65d60bd6a084f5fb8a03 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0019-9800000000-5cfef19dd8c8707bef7a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00yi-0179000000-d117b299438bee3a1a40 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uk9-0592000000-db6f5abfab26c98d8af4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0wbc-5960000000-915e1f050484eedb63d2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-2492000000-1208b9128bdb1fd8688e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0h90-7984000000-2636a4af2a05389c39e5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pb9-7910000000-dce1bda7d8788ebd90af | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003r-1169000000-37af2930ba0a18fdfe2d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bu3-9317000000-05676e3bb792b14a9ea9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9410000000-9f5dbeeb51b744740d72 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f8a-0659000000-98f0c5a2f817d77728ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uk9-0951000000-f475151e03af91aeeac3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006w-9410000000-abd7ef88c6f0a3d44980 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Dekany, Gyula; Agoston, Karoly; Bajza, Istvan; Boejstrup, Marie; Kroeger, Lars. Process for the large-scale preparation of N-acetyllactosamine, lactosamine, lactosamine salts and lactosamine-containing oligosaccharides. PCT Int. Appl. (2007), 44pp. |
|---|