| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:50:11 UTC |
|---|
| Update Date | 2020-05-21 16:29:01 UTC |
|---|
| BMDB ID | BMDB0002031 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Ureidoisobutyric acid |
|---|
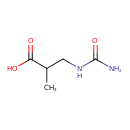
| Description | Ureidoisobutyric acid, also known as 3-ureidoisobutyrate or beta-uba, belongs to the class of organic compounds known as ureas. Ureas are compounds containing two amine groups joined by a carbonyl (C=O) functional group. Ureidoisobutyric acid exists in all living organisms, ranging from bacteria to humans. Based on a literature review a small amount of articles have been published on Ureidoisobutyric acid. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-((Aminocarbonyl)amino)-2-methylpropanoic acid | ChEBI | | 3-Ureidoisobutyrate | ChEBI | | beta-UBA | ChEBI | | beta-Ureidoisobutyric acid | ChEBI | | 3-((Aminocarbonyl)amino)-2-methylpropanoate | Generator | | 3-Ureidoisobutyric acid | Generator | | b-UBA | Generator | | Β-uba | Generator | | b-Ureidoisobutyrate | Generator | | b-Ureidoisobutyric acid | Generator | | beta-Ureidoisobutyrate | Generator | | Β-ureidoisobutyrate | Generator | | Β-ureidoisobutyric acid | Generator | | Ureidoisobutyrate | Generator | | 3-carbamoylamino-2-Methylpropanoate | HMDB | | 3-carbamoylamino-2-Methylpropanoic acid | HMDB | | DL-beta-Ureidoisobutyrate | HMDB | | DL-beta-Ureidoisobutyric acid | HMDB | | N-Carbamyl-amino isobutyrate | HMDB | | N-Carbamyl-b-aminoisobutyric acid | HMDB | | N-Carbamyl-beta-aminoisobutyric acid | HMDB |
|
|---|
| Chemical Formula | C5H10N2O3 |
|---|
| Average Molecular Weight | 146.1445 |
|---|
| Monoisotopic Molecular Weight | 146.069142196 |
|---|
| IUPAC Name | 3-(carbamoylamino)-2-methylpropanoic acid |
|---|
| Traditional Name | β-ureidoisobutyric acid |
|---|
| CAS Registry Number | 2905-86-4 |
|---|
| SMILES | CC(CNC(N)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H10N2O3/c1-3(4(8)9)2-7-5(6)10/h3H,2H2,1H3,(H,8,9)(H3,6,7,10) |
|---|
| InChI Key | PHENTZNALBMCQD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as ureas. Ureas are compounds containing two amine groups joined by a carbonyl (C=O) functional group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic carbonic acids and derivatives |
|---|
| Sub Class | Ureas |
|---|
| Direct Parent | Ureas |
|---|
| Alternative Parents | |
|---|
| Substituents | - Urea
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-006x-9100000000-5fb552346f153d8b9141 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00xr-9510000000-6396a4042b792437e39a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ug1-4900000000-57fe49b6570a3a56075e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9400000000-d75bc53f6bd893972265 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-505e1e587ff8e3f47c4b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udj-6900000000-d0e112adeac57b5cc676 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zfu-9600000000-38cc35413fc058ff2f4f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-46efbc5db1bc9c62db04 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-8900000000-51286b64a41bad89fd80 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05gi-9100000000-835c9b02eecbd89baac8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-40f2b4e3a071943603ee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-3900000000-9dfc177176195ee47192 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052f-9000000000-0f56471f86b7f2230673 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-60a7db0ec3c9213e9347 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|