| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:51:16 UTC |
|---|
| Update Date | 2020-05-21 16:26:55 UTC |
|---|
| BMDB ID | BMDB0002104 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
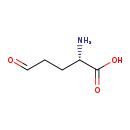
| Common Name | L-Glutamic-gamma-semialdehyde |
|---|
| Description | L-Glutamic gamma-semialdehyde, also known as L-glutamate 5-semialdehyde or 5-oxo-L-norvaline, belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. L-Glutamic gamma-semialdehyde is a very strong basic compound (based on its pKa). L-Glutamic gamma-semialdehyde exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-oxo-L-Norvaline | ChEBI | | L-Glutamate 5-semialdehyde | ChEBI | | L-Glutamate gamma-semialdehyde | ChEBI | | L-Glutamic acid 5-semialdehyde | Generator | | L-Glutamate g-semialdehyde | Generator | | L-Glutamate γ-semialdehyde | Generator | | L-Glutamic acid g-semialdehyde | Generator | | L-Glutamic acid gamma-semialdehyde | Generator | | L-Glutamic acid γ-semialdehyde | Generator | | L-Glutamic g-semialdehyde | Generator | | L-Glutamic γ-semialdehyde | Generator | | Glutamate-semialdehyde | HMDB | | Glutamic gamma-semialdehyde | HMDB | | L-Glutamate-5-semialdehyde | HMDB | | L-Glutamate-gamma-semialdehyde | HMDB | | L-Glutamic-gamma-semialdehyde | HMDB | | gamma-Glutamyl semialdehyde | HMDB | | Glutamate gamma-semialdehyde | HMDB | | Glutamic acid gamma-semialdehyde | HMDB | | Glutamic acid gamma-semialdehyde, (L)-isomer | HMDB |
|
|---|
| Chemical Formula | C5H9NO3 |
|---|
| Average Molecular Weight | 131.1299 |
|---|
| Monoisotopic Molecular Weight | 131.058243159 |
|---|
| IUPAC Name | (2S)-2-amino-5-oxopentanoic acid |
|---|
| Traditional Name | 4-carboxy-4-aminobutanal |
|---|
| CAS Registry Number | 496-92-4 |
|---|
| SMILES | N[C@@H](CCC=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H9NO3/c6-4(5(8)9)2-1-3-7/h3-4H,1-2,6H2,(H,8,9)/t4-/m0/s1 |
|---|
| InChI Key | KABXUUFDPUOJMW-BYPYZUCNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Fatty acid
- Alpha-hydrogen aldehyde
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Aldehyde
- Hydrocarbon derivative
- Organic oxide
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Amine
- Organic nitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-059l-9000000000-c907c5e28c6d3b047532 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-000i-9200000000-b09702360bf061bd64db | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-02a9-9700000000-ffc8c81a881bac7a0ca7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-9100000000-7061d8b19402303b61af | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-fa50d608bfaec666722d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-1900000000-485c1c464e6d065cb4ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01q9-7900000000-cd2ef70374a324d28ee0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-8685f6b3a1d6936c24c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-30a10f2c9d729eb39f1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03e9-6900000000-22f02534479a7a01c5bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-cb0e34b8ef2d18870f9b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9200000000-69607aaecb41dd2d014a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014u-9000000000-d6928830fd6c484b658f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-9000000000-915ef2dc6a2dda8f2ee6 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|