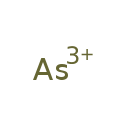| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:53:42 UTC |
|---|
| Update Date | 2020-06-04 19:27:15 UTC |
|---|
| BMDB ID | BMDB0002290 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Arsenic |
|---|
| Description | Arsenic, also known as arsen or as(3+), belongs to the class of inorganic compounds known as homogeneous metalloid compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a metalloid atom. Arsenic exists as a solid, possibly soluble (in water), and possibly neutral molecule. Arsenic is formally rated as a carcinogen (by IARC 1) and is also a potentially toxic compound. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| As(3+) | ChEBI | | Arsen | HMDB | | Arsenic black | HMDB | | Arsenic elemental | HMDB | | As | HMDB | | Grey arsenic | HMDB | | Metallic arsenic | HMDB |
|
|---|
| Chemical Formula | As |
|---|
| Average Molecular Weight | 74.9216 |
|---|
| Monoisotopic Molecular Weight | 74.921596417 |
|---|
| IUPAC Name | arsenic(3+) ion |
|---|
| Traditional Name | arsenic(3+) ion |
|---|
| CAS Registry Number | 7440-38-2 |
|---|
| SMILES | [As+3] |
|---|
| InChI Identifier | InChI=1S/As/q+3 |
|---|
| InChI Key | LULLIKNODDLMDQ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of inorganic compounds known as homogeneous metalloid compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a metalloid atom. |
|---|
| Kingdom | Inorganic compounds |
|---|
| Super Class | Homogeneous metal compounds |
|---|
| Class | Homogeneous metalloid compounds |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Homogeneous metalloid compounds |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | |
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | > 615 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | |
|---|
| Biological Properties |
|---|
| Cellular Locations | Not Available |
|---|
| Biospecimen Locations | |
|---|
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Milk | Detected and Quantified | 0.387 - 0.467 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.11 - 0.18 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.13 +/- 0.01 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.14 +/- 0.01 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.14 +/- 0.01 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.11 +/- 0.05 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.0525 +/- 0.000801 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.0547 +/- 0.00133 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.0575 +/- 0.00120 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.0467 +/- 0.00133 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.0280 +/- 0.00133 uM | Not Specified | Not Specified | Normal | | details | | Milk | Detected and Quantified | 0.0440 +/- 0.00267 uM | Not Specified | Not Specified | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0002290 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB003763 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 94549 |
|---|
| KEGG Compound ID | C06269 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Arsenic |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 104734 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 35828 |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| General References | - T Rana, S Sarkar, T Mandal, K Bhattacharyya, A Roy, L Kol (2017). Tanmoy Rana et al. Contribution of arsenic from agricultural food chain to cow milk in highly arsenic prone zone in Nadia District of West Bengal in India.The Internet Journal of Veterinary Medicine Vol 4(2). The Internet Journal of Veterinary Medicine .
- Z. Dobrzański, R. Kołacz, H. Górecka, K. Chojnacka, A. Bartkowiak. (2005). Z. Dobrzański, R. Kołacz, H. Górecka, K. Chojnacka, A. Bartkowiak. 2005. The Content of Microelements and Trace Elements in Raw Milk from Cows in the Silesian Region. Pol. J. Environ. Stud. 14(5):685–689. Polish Journal of Environmental Studies.
- A. Foroutan et al. (2019). A. Foroutan et al. The Chemical Composition of Commercial Cow's Milk (in preparation). Journal of Agricultural and Food Chemistry.
- Patricia Cava-Montesinos, M. Luisa Cervera Agustín Pastor Miguel de la Guardia (2005). Patricia Cava-Montesinos, M. Luisa Cervera Agustín Pastor Miguel de la Guardia. 2005. Room temperature acid sonication ICP-MS multielemental analysis of milk.Analytica Chimica Acta Volume 531, Issue 1, Pages 111-123. Analytica Chimica Acta.
|
|---|