| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:03:58 UTC |
|---|
| Update Date | 2020-04-22 15:12:05 UTC |
|---|
| BMDB ID | BMDB0003361 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
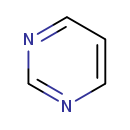
| Common Name | Pyrimidine |
|---|
| Description | Pyrimidine, also known as pyrimidine base or 1,3-diazin, belongs to the class of organic compounds known as pyrimidines and pyrimidine derivatives. Pyrimidines and pyrimidine derivatives are compounds containing a pyrimidne ring, which is a six-member aromatic heterocycle which consists of two nitrogen atoms (at positions 1 and 3) and four carbon atoms. Pyrimidine exists as a solid, possibly soluble (in water), and a moderately basic compound (based on its pKa) molecule. Pyrimidine exists in all living organisms, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,3-Diazin | ChEBI | | 1,3-Diazine | ChEBI | | m-Diazine | ChEBI | | Metadiazine | ChEBI | | Pyrimidin | ChEBI | | Pyrimidine base | ChEBI | | 1,3-Diazabenzene | HMDB | | Miazine | HMDB | | PY | HMDB | | PYR | HMDB | | Pyrimidine dimer | HMDB |
|
|---|
| Chemical Formula | C4H4N2 |
|---|
| Average Molecular Weight | 80.088 |
|---|
| Monoisotopic Molecular Weight | 80.037448138 |
|---|
| IUPAC Name | pyrimidine |
|---|
| Traditional Name | pyrimidine |
|---|
| CAS Registry Number | 25247-63-6 |
|---|
| SMILES | C1=CN=CN=C1 |
|---|
| InChI Identifier | InChI=1S/C4H4N2/c1-2-5-4-6-3-1/h1-4H |
|---|
| InChI Key | CZPWVGJYEJSRLH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidines and pyrimidine derivatives. Pyrimidines and pyrimidine derivatives are compounds containing a pyrimidne ring, which is a six-member aromatic heterocycle which consists of two nitrogen atoms (at positions 1 and 3) and four carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Diazines |
|---|
| Sub Class | Pyrimidines and pyrimidine derivatives |
|---|
| Direct Parent | Pyrimidines and pyrimidine derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine
- Heteroaromatic compound
- Azacycle
- Organic nitrogen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 22 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000 mg/mL at 25 °C | Not Available | | LogP | -0.4 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-9000000000-d94930210c167d44af20 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-9000000000-bcbcbca1a7f4d54af095 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0udi-9000000000-9589f26615aa6ffdf19f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9000000000-d6fe4ae4575881938f2c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-001i-9000000000-036ccbc8350c743f75f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9000000000-e6219273709c557c3698 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f89-9000000000-f3e79d098ad4304a8384 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udr-9000000000-fcf5f26e0c97b91147ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9000000000-8fe843c91e37282a20a3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-25f44ab9caf4fde752e0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-9000000000-8f30bec7099c7d8746ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9000000000-a836173ec317f96549bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-9000000000-1371ace385ac68f9f317 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-1573e0ce56775ede12c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9000000000-90831ac2a79e645518ba | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-90831ac2a79e645518ba | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-f1f0ba91ed28b4268b72 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Pan L, Yu J, Mi Z, Mo L, Jin H, Yao C, Ren D, Menghe B: A Metabolomics Approach Uncovers Differences between Traditional and Commercial Dairy Products in Buryatia (Russian Federation). Molecules. 2018 Mar 22;23(4). pii: molecules23040735. doi: 10.3390/molecules23040735. [PubMed:29565828 ]
|
|---|