| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:08:36 UTC |
|---|
| Update Date | 2020-05-21 16:28:31 UTC |
|---|
| BMDB ID | BMDB0004225 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 2-Oxoarginine |
|---|
| Description | 2-Oxoarginine belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. 2-Oxoarginine is possibly soluble (in water) and a very strong basic compound (based on its pKa). 2-Oxoarginine can be biosynthesized from D-arginine through the action of the enzyme D-amino-acid oxidase. In cattle, 2-oxoarginine is involved in the metabolic pathway called the D-arginine and D-ornithine metabolism pathway. 2-Oxoarginine is a potentially toxic compound. |
|---|
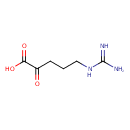
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-oxo-5-Guanidino-pentanoate | ChEBI | | 2-oxo-5-Guanidinopentanoate | ChEBI | | 2-oxo-5-Guanidinovaleric acid | ChEBI | | 5-((Aminoiminomethyl)amino)-2-oxopentanoic acid | ChEBI | | 5-Guanidino-2-oxo-pentanoate | ChEBI | | 5-Guanidino-2-oxopentanoate | ChEBI | | alpha-Keto-delta-guanidinopentanoic acid | ChEBI | | alpha-Keto-delta-guanidinovaleric acid | ChEBI | | 2-oxo-5-Guanidino-pentanoic acid | Generator | | 2-oxo-5-Guanidinopentanoic acid | Generator | | 2-oxo-5-Guanidinovalerate | Generator | | 5-((Aminoiminomethyl)amino)-2-oxopentanoate | Generator | | 5-Guanidino-2-oxo-pentanoic acid | Generator | | 5-Guanidino-2-oxopentanoic acid | Generator | | a-Keto-delta-guanidinopentanoate | Generator | | a-Keto-delta-guanidinopentanoic acid | Generator | | alpha-Keto-delta-guanidinopentanoate | Generator | | Α-keto-δ-guanidinopentanoate | Generator | | Α-keto-δ-guanidinopentanoic acid | Generator | | a-Keto-delta-guanidinovalerate | Generator | | a-Keto-delta-guanidinovaleric acid | Generator | | alpha-Keto-delta-guanidinovalerate | Generator | | Α-keto-δ-guanidinovalerate | Generator | | Α-keto-δ-guanidinovaleric acid | Generator | | a-Keto-δ-guanidinovalerate | HMDB | | a-Keto-δ-guanidinovaleric acid | HMDB | | a-Keto-δ-guanidinopentanoate | HMDB | | a-Keto-δ-guanidinopentanoic acid | HMDB | | 5-(Diaminomethylideneamino)-2-oxopentanoate | HMDB | | 5-(Diaminomethylideneamino)-2-oxopentanoic acid | HMDB | | 5-Guanidino-2-oxovaleric acid | HMDB | | a-Keto-D-guanidinovaleric acid | HMDB | | D-Guanidino-a-oxovaleric acid | HMDB | | D-Guanido-a-ketovaleric acid | HMDB | | delta-Guanidino-alpha-oxovaleric acid | HMDB | | delta-Guanido-alpha-ketovaleric acid | HMDB | | 2-Oxoarginine | ChEBI | | 5-[(Aminoiminomethyl)amino]-2-oxopentanoic acid | PhytoBank | | δ-Guanidino-α-oxovaleric acid | PhytoBank | | δ-Guanido-α-ketovaleric acid | PhytoBank |
|
|---|
| Chemical Formula | C6H11N3O3 |
|---|
| Average Molecular Weight | 173.1698 |
|---|
| Monoisotopic Molecular Weight | 173.080041233 |
|---|
| IUPAC Name | 5-[(diaminomethylidene)amino]-2-oxopentanoic acid |
|---|
| Traditional Name | 5-[(diaminomethylidene)amino]-2-oxopentanoic acid |
|---|
| CAS Registry Number | 3715-10-4 |
|---|
| SMILES | NC(N)=NCCCC(=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H11N3O3/c7-6(8)9-3-1-2-4(10)5(11)12/h1-3H2,(H,11,12)(H4,7,8,9) |
|---|
| InChI Key | ARBHXJXXVVHMET-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Short-chain keto acids and derivatives |
|---|
| Direct Parent | Short-chain keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain keto acid
- Alpha-keto acid
- Alpha-hydroxy ketone
- Guanidine
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Carboximidamide
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Membrane
- Peroxisome
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0096-9100000000-303513b0b209bca56206 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00r6-9710000000-95bf9765fd1fa4c2cd82 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-0900000000-4c337e4db09d448ecc63 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0563-9600000000-d97800b4db5ae44aebba | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9200000000-6286bbf95e4a4716e7d4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-2900000000-67d1c875d2da6e075630 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9800000000-1a3c6500b4d652bcaadb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-89a2ffbc759f6a2a5922 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-2900000000-48efdecf4dbd7823ddff | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08fu-6900000000-0ca535d235f66409d225 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-9000000000-82063658464a6dc17369 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ab9-1900000000-05cd70e6e18e08b35be8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0900-7900000000-aec8a82d0097632e2cc6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9000000000-c27deb914dc894c12d6c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-2900000000-fcee1bc9a64cc54c8a61 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014m-9100000000-884cb4184447c0cb50bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ox-9000000000-d71574817dabeaf6e5e0 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|