| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:13:32 UTC |
|---|
| Update Date | 2020-05-11 20:57:18 UTC |
|---|
| BMDB ID | BMDB0004947 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
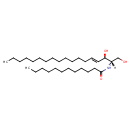
| Common Name | Ceramide (d18:1/12:0) |
|---|
| Description | Ceramide (d18:1/12:0), also known as N-(lauroyl)sphing-4-enine or N-(dodecanoyl)ceramide, belongs to the class of organic compounds known as ceramides. These are lipid molecules containing a sphingosine in which the amine group is linked to a fatty acid. Thus, ceramide (D18:1/12:0) is considered to be a ceramide lipid molecule. Ceramide (d18:1/12:0) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Cer(d18:1/12:0) | ChEBI | | Dodecyl sphingosine | ChEBI | | Laurylsphingosine | ChEBI | | N-(Dodecanoyl)ceramide | ChEBI | | N-(Dodecanoyl)sphing-4-enine | ChEBI | | N-(Lauroyl)ceramide | ChEBI | | N-(Lauroyl)sphing-4-enine | ChEBI | | N-Dodecanoylsphing-4-enine | ChEBI | | N-Lauroylsphing-4-enine | ChEBI | | N-Lauroylsphingosine | ChEBI | | (2S,3R,4E)-2-acylamino-1,3-Octadec-4-enediol | HMDB | | (2S,3R,4E)-2-Acylaminooctadec-4-ene-1,3-diol | HMDB | | Cer | HMDB | | Ceramide | HMDB | | N-Acylsphingosine | HMDB | | N-[(1S,2R,3E)-2-Hydroxy-1-(hydroxymethyl)-3-heptadecenyl]-octadecanamide | HMDB | | C12-Ceramide | HMDB | | Cer d18:1/12:0 | HMDB | | Ceramide (d18:1,C12:0) | HMDB | | Ceramide (d18:1/12:0) | HMDB | | Ceramide 12 | HMDB | | Ceramide(d18:1/12:0) | HMDB | | N-Lauroyl-D-erythro-sphingosine | HMDB | | N-Laurylsphingosine | HMDB |
|
|---|
| Chemical Formula | C30H59NO3 |
|---|
| Average Molecular Weight | 481.7944 |
|---|
| Monoisotopic Molecular Weight | 481.449494759 |
|---|
| IUPAC Name | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]dodecanamide |
|---|
| Traditional Name | N-(dodecanoyl)-ceramide |
|---|
| CAS Registry Number | 74713-60-3 |
|---|
| SMILES | CCCCCCCCCCCCC\C=C\[C@@H](O)[C@H](CO)NC(=O)CCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C30H59NO3/c1-3-5-7-9-11-13-14-15-16-18-19-21-23-25-29(33)28(27-32)31-30(34)26-24-22-20-17-12-10-8-6-4-2/h23,25,28-29,32-33H,3-22,24,26-27H2,1-2H3,(H,31,34)/b25-23+/t28-,29+/m0/s1 |
|---|
| InChI Key | HXFPPRPLRSPNIB-VARSQMIESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as ceramides. These are lipid molecules containing a sphingosine in which the amine group is linked to a fatty acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Sphingolipids |
|---|
| Sub Class | Ceramides |
|---|
| Direct Parent | Ceramides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Ceramide
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Carboxamide group
- Secondary alcohol
- Secondary carboxylic acid amide
- Carboxylic acid derivative
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Endosome
- Intracellular membrane
- Membrane
- Mitochondria
- Myelin sheath
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Insoluble | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-022i-5785598000-7031136168734e1ef5ee | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-2090000000-ae80000c40a296a1c5d8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-2090000000-a13bd310025f03a29721 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-2090000000-8d5084b0da3675b3f3f3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-2090000000-d4824df02fcc2092417c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-2090000000-f919f09dc660be85f13e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000900000-fe0f573db63b1b90fd2f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-0050900000-612b7fe1fb533df6232b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03e9-0090600000-9abebdbdb27db5a25cb2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000900000-fa0543ff22d51d18b573 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0010900000-3de9968cb166260af69f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001k-0220900000-1f62f1430f619c4887d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000900000-2b1019f1dfd77202b1a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000900000-2b1019f1dfd77202b1a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-0000900000-aa4c78e902645a2bb092 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000900000-5acec5af0818f408e044 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-0050900000-f3ad47a5043cb0aa6c70 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03e9-0090600000-797db203768da46183b0 | View in MoNA |
|---|
|
|---|