| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:13:34 UTC |
|---|
| Update Date | 2020-05-11 20:57:19 UTC |
|---|
| BMDB ID | BMDB0004948 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Ceramide (d18:1/9Z-18:1) |
|---|
| Description | Ceramide (d18:1/9Z-18:1), also known as C18:1 cer or N-(9Z-octadecenoyl)-ceramide, belongs to the class of organic compounds known as long-chain ceramides. These are ceramides bearing a long chain fatty acid. Thus, ceramide (D18:1/9Z-18:1) is considered to be a ceramide lipid molecule. Ceramide (d18:1/9Z-18:1) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
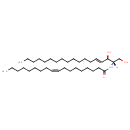
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| C18:1 Cer | ChEBI | | Cer(d18:1/18:1(9Z)) | ChEBI | | N-(9Z-Octadecenoyl)-ceramide | ChEBI | | N-(9Z-Octadecenoyl)-sphing-4-enine | ChEBI | | N-(Oleoyl)-ceramide | ChEBI | | N-[(9Z)-Octadecenoyl]sphing-4-enine | ChEBI | | N-[(9Z)-Octadecenoyl]sphingosine | ChEBI | | N-Oleoylsphing-4-enine | ChEBI | | (2S,3R,4E)-2-acylamino-1,3-Octadec-4-enediol | HMDB | | (2S,3R,4E)-2-Acylaminooctadec-4-ene-1,3-diol | HMDB | | Cer | HMDB | | Ceramide | HMDB | | N-Acylsphingosine | HMDB | | N-[(1S,2R,3E)-2-Hydroxy-1-(hydroxymethyl)-3-heptadecenyl]-octadecanamide | HMDB | | N-Oleoylsphingosine | MeSH, HMDB | | Oleoylsphingosine | MeSH, HMDB | | C18:1-Ceramide | HMDB | | Cer d18:1/18:1 | HMDB | | Cer d18:1/18:1(9Z) | HMDB | | Cer(d18:1/18:1) | HMDB | | Ceramide (d18:1,C18:1(9Z)) | HMDB | | Ceramide (d18:1/18:1(9Z)) | HMDB | | Ceramide (d18:1/18:1) | HMDB | | Ceramide (d18:1/9Z-18:1) | HMDB | | Ceramide 18:1 | HMDB | | Ceramide(d18:1/18:1(9Z)) | HMDB | | Ceramide(d18:1/18:1) | HMDB | | Ceramide(d18:1/9Z-18:1) | HMDB | | D-erythro-1,3-Dihydroxy-2-(cis-9-octadecenoylamido)-trans-4-octadecene | HMDB | | N-Oleoyl-4-sphingenine | HMDB | | N-Oleoyl-D-sphingosine | HMDB |
|
|---|
| Chemical Formula | C36H69NO3 |
|---|
| Average Molecular Weight | 563.938 |
|---|
| Monoisotopic Molecular Weight | 563.527745079 |
|---|
| IUPAC Name | (9Z)-N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]octadec-9-enamide |
|---|
| Traditional Name | N-(oleoyl)-ceramide |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CCCCCCCCCCCCC\C=C\[C@@H](O)[C@H](CO)NC(=O)CCCCCCC\C=C/CCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C36H69NO3/c1-3-5-7-9-11-13-15-17-18-20-22-24-26-28-30-32-36(40)37-34(33-38)35(39)31-29-27-25-23-21-19-16-14-12-10-8-6-4-2/h17-18,29,31,34-35,38-39H,3-16,19-28,30,32-33H2,1-2H3,(H,37,40)/b18-17-,31-29+/t34-,35+/m0/s1 |
|---|
| InChI Key | OBFSLMQLPNKVRW-RHPAUOISSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as long-chain ceramides. These are ceramides bearing a long chain fatty acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Sphingolipids |
|---|
| Sub Class | Ceramides |
|---|
| Direct Parent | Long-chain ceramides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Long-chain ceramide
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Carboxamide group
- Secondary alcohol
- Secondary carboxylic acid amide
- Carboxylic acid derivative
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Endosome
- Intracellular membrane
- Membrane
- Mitochondria
- Myelin sheath
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Insoluble | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0229-5438589000-b23a5b0a9070bb8f69fc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000090000-42bbdc6df2df47887c98 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0050090000-5f063501ce7556661965 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03ea-0090060000-fb661b5553f476ecd13b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000090000-356968936fbf32f32cf6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0050090000-72946d4294c83854c648 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03ea-0090060000-3c7f6c443aba753b1f19 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000090000-5d8eebdcf3a4579fd637 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0000090000-5d8eebdcf3a4579fd637 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0000090000-3d1ba9f1a6f4cc3007c0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0000090000-08981d9b789f077be231 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0010090000-9c2bedfc097f18a9ba61 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03f0-0030090000-0319ae7a5e287b6ed17c | View in MoNA |
|---|
|
|---|