| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:13:35 UTC |
|---|
| Update Date | 2020-05-11 20:57:20 UTC |
|---|
| BMDB ID | BMDB0004949 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Ceramide (d18:1/16:0) |
|---|
| Description | Cer(d18:1/16:0), also known as N-hexadecanoyl-sphing-4-enine, is a ceramide (Cer). Ceramides are members of the class of compounds known as sphingolipids (SPs), or glycosylceramides. SPs are lipids containing a backbone of sphingoid bases (e.g. sphingosine or sphinganine) that are often covalently bound to a fatty acid derivative through N-acylation. SPs are found in cell membranes, particularly in peripheral nerve cells and the cells found in the central nervous system (including the brain and spinal cord). Sphingolipids are extremely versatile molecules that have functions controlling fundamental cellular processes such as cell division, differentiation, and cell death. Impairments associated with sphingolipid metabolism are associated with many common human diseases such as diabetes, various cancers, microbial infections, diseases of the cardiovascular and respiratory systems, Alzheimer’s disease and other neurological syndromes. The biosynthesis and catabolism of sphingolipids involves a large number of intermediate metabolites where many different enzymes are involved. Simple sphingolipids, which include the sphingoid bases and ceramides, make up the early products of the sphingolipid synthetic pathways, while complex sphingolipids may be formed by the addition of head groups to the ceramide template (Wikipedia). In humans, ceramides are phosphorylated to ceramide phosphates (CerPs) through the action of a specific ceramide kinase (CerK). Ceramide phosphates are important metabolites of ceramides as they act as a mediators of the inflammatory response. Ceramides are also one of the hydrolysis byproducts of sphingomyelins (SMs) through the action of the enzyme sphingomyelin phosphodiesterase, which has been identified in the subcellular fractions of human epidermis (PMID: 25935 ) and many other tissues. Ceramides can also be synthesized from serine and palmitate in a de novo pathway and are regarded as important cellular signals for inducing apoptosis (PMID: 14998372 ). Ceramides are key in the biosynthesis of glycosphingolipids and gangliosides. In terms of its appearance and structure, Cer(d18:1/16:0) is a colorless solid that consists of an unsaturated 18-carbon sphingoid base with an attached saturated hexadecanoyl fatty acid side chain. In most mammalian SPs, the 18-carbon sphingoid bases are predominant (PMID: 9759481 ). |
|---|
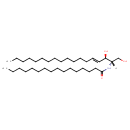
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S,3R,4E)-2-Acylamino-1,3-octadec-4-enediol | ChEBI | | (2S,3R,4E)-2-Acylaminooctadec-4-ene-1,3-diol | ChEBI | | C16 Cer | ChEBI | | C16 Ceramide | ChEBI | | C16-0(Palmitoyl)ceramide | ChEBI | | Ceramide (D18:1/16:0) | ChEBI | | N-(Hexadecanoyl)-sphing-4-enine | ChEBI | | N-(Hexadecanoyl)ceramide | ChEBI | | N-(Hexadecanoyl)sphing-4-enine | ChEBI | | N-(Palmitoyl)ceramide | ChEBI | | N-Hexadecanoyl-D-erythro-sphingosine | ChEBI | | N-Hexadecanoylsphing-4-enine | ChEBI | | N-Palmitoyl-sphingosine | ChEBI | | N-Palmitoylsphing-4-enine | ChEBI | | N-Palmitoylsphingosine | ChEBI | | Cer | HMDB | | Ceramide | HMDB | | N-Acylsphingosine | HMDB | | N-[(1S,2R,3E)-2-Hydroxy-1-(hydroxymethyl)-3-heptadecenyl]-octadecanamide | HMDB | | C16-Ceramide | HMDB | | NFA(C16)cer | HMDB | | Palmitoylceramide | HMDB | | C(16)-Ceramide | HMDB | | N-Palmitoylsphingosine, (r*,s*-(e))-(+-) | HMDB | | C16-Palmitoylceramide | HMDB | | N-Palmitoylsphingosine, R-(r*,s*-(e)) | HMDB | | PCer CPD | HMDB | | Ceramide-C16 | HMDB | | (2S,3R,4E)-2-N-Palmitoyloctadecasphinga-4-ene | HMDB | | C16:0-Ceramide | HMDB | | Ceramide (D18:1,C16:0) | HMDB | | Ceramide 16 | HMDB | | Ceramide(D18:1/16:0) | HMDB | | D-Erythro-C16-ceramide | HMDB | | D-Erythro-N-hexadecanoylsphingenine | HMDB | | D-Erythro-N-palmitoylsphingosine | HMDB | | D-Erythro-delta4-ceramide | HMDB | | D-Erythro-δ4-ceramide | HMDB | | N-Hexadecanoyl-D-erythro-ceramide | HMDB | | N-Hexadecanoylsphingosine | HMDB | | N-Palmitoyl 4-sphingenine | HMDB | | N-Palmitoyl ceramide | HMDB | | N-Palmitoyl-D-erythro-sphingosine | HMDB | | N-Palmitoyl-D-sphingosine | HMDB | | N-(Hexadecanoyl)-sphingosine | MetBuilder | | N-(Hexadecanoyl)-D-erythro-sphingosine | MetBuilder | | N-(Hexadecanoyl)-4-sphingenine | MetBuilder | | N-(Hexadecanoyl)-D-sphingosine | MetBuilder | | N-(Hexadecanoyl)-sphingenine | MetBuilder | | N-(Hexadecanoyl)-erythro-4-sphingenine | MetBuilder | | Cer(D18:1/16:0) | ChEBI |
|
|---|
| Chemical Formula | C34H67NO3 |
|---|
| Average Molecular Weight | 537.9007 |
|---|
| Monoisotopic Molecular Weight | 537.512095015 |
|---|
| IUPAC Name | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]hexadecanamide |
|---|
| Traditional Name | N-(palmitoyl)-ceramide |
|---|
| CAS Registry Number | 4201-58-5 |
|---|
| SMILES | CCCCCCCCCCCCCCCC(=O)N[C@@H](CO)[C@H](O)\C=C\CCCCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C34H67NO3/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-33(37)32(31-36)35-34(38)30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h27,29,32-33,36-37H,3-26,28,30-31H2,1-2H3,(H,35,38)/b29-27+/t32-,33+/m0/s1 |
|---|
| InChI Key | YDNKGFDKKRUKPY-TURZORIXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as long-chain ceramides. These are ceramides bearing a long chain fatty acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Sphingolipids |
|---|
| Sub Class | Ceramides |
|---|
| Direct Parent | Long-chain ceramides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Long-chain ceramide
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Carboxamide group
- Secondary alcohol
- Secondary carboxylic acid amide
- Carboxylic acid derivative
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Endosome
- Intracellular membrane
- Membrane
- Mitochondria
- Myelin sheath
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Insoluble | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00y0-4371679000-4350654880fad493e81d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0090020000-a4c08ce2d2d5bc07212c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0090020000-6b6cc500a909ad02bd59 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0090020000-4a672523b98af8ff8c7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000090000-f294f0ced8f149249815 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-0050090000-80f4b14827e42ec58fea | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03l0-0090050000-c9f32bf831226119babd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000090000-876883608819d763f04e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-0050090000-22ff2434faf08fcef599 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03l0-0090050000-e6a7f6704ac5e854065c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000090000-15b6716e7356e06043b6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0010090000-e14e9c0c2e28b7a12b19 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ue0-0040090000-a44d6a95bf21d0975da6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000090000-d2d11b266d3c5c7e73ff | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-0000090000-d2d11b266d3c5c7e73ff | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-0000190000-6ce0a8c6d5140383d837 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|