| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:21:06 UTC |
|---|
| Update Date | 2020-05-21 16:28:31 UTC |
|---|
| BMDB ID | BMDB0006272 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5-Amino-2-oxopentanoic acid |
|---|
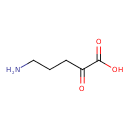
| Description | 5-Amino-2-oxopentanoic acid, also known as 5-amino-2-oxopentanoic acid or 5-amino-2-oxopentanoic acid, belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. 5-Amino-2-oxopentanoic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. 5-Amino-2-oxopentanoic acid exists in all living organisms, ranging from bacteria to humans. 5-Amino-2-oxopentanoic acid can be biosynthesized from D-ornithine; which is mediated by the enzyme D-amino-acid oxidase. In cattle, 5-amino-2-oxopentanoic acid is involved in the metabolic pathway called the D-arginine and D-ornithine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-oxo-5-Amino-pentanoate | ChEBI | | 2-oxo-5-Aminopentanoate | ChEBI | | 2-oxo-5-Aminovalerate | ChEBI | | 5-Amino-2-oxopentanoate | ChEBI | | 5-Amino-2-oxovaleric acid | ChEBI | | alpha-Keto-delta-aminopentanoate | ChEBI | | 2-oxo-5-Amino-pentanoic acid | Generator | | 2-oxo-5-Aminopentanoic acid | Generator | | 2-oxo-5-Aminovaleric acid | Generator | | 5-Amino-2-oxovalerate | Generator | | a-Keto-delta-aminopentanoate | Generator | | a-Keto-delta-aminopentanoic acid | Generator | | alpha-Keto-delta-aminopentanoic acid | Generator | | Α-keto-δ-aminopentanoate | Generator | | Α-keto-δ-aminopentanoic acid | Generator | | a-Keto-δ-aminopentanoate | HMDB | | a-Keto-δ-aminopentanoic acid | HMDB |
|
|---|
| Chemical Formula | C5H9NO3 |
|---|
| Average Molecular Weight | 131.1299 |
|---|
| Monoisotopic Molecular Weight | 131.058243159 |
|---|
| IUPAC Name | 5-amino-2-oxopentanoic acid |
|---|
| Traditional Name | 5-amino-2-oxopentanoic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NCCCC(=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H9NO3/c6-3-1-2-4(7)5(8)9/h1-3,6H2,(H,8,9) |
|---|
| InChI Key | BWHGMFYTDQEALD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Short-chain keto acids and derivatives |
|---|
| Direct Parent | Short-chain keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain keto acid
- Alpha-keto acid
- Gamma-aminoketone
- Alpha-hydroxy ketone
- Amino acid or derivatives
- Amino acid
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organooxygen compound
- Primary amine
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Amine
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Membrane
- Peroxisome
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-9000000000-4f2ab9a8ceda96ab2b3d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0fk9-9600000000-ff46e8c1649cc828f7b0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-02u1-4900000000-e6f994d6e834039502b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02td-9300000000-e8e8edb03b7aaff8ee8e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-5b1e326206ccaec9c5d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-3900000000-680079416681a6d56931 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01qi-9500000000-56702f30bf4675c2b75a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05nf-9000000000-7a44c7ada571fc80c95b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9200000000-b31fe2b03dd6d43aaf76 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01bd-9000000000-ca1b69887ff2e7d07f6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-9000000000-f6e5fdeca83cd716bca7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-223ace0a9362994292ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9200000000-72f5d31ad31e1076f60b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-9000000000-6915b5779a08500cd58e | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|