| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:22:02 UTC |
|---|
| Update Date | 2020-05-21 16:28:59 UTC |
|---|
| BMDB ID | BMDB0006454 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | L-2-Amino-3-oxobutanoic acid |
|---|
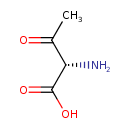
| Description | L-2-Amino-3-oxobutanoic acid, also known as (S)-L-L-l-2-amino-3-oxobutanoic acid or L-L-l-2-amino-3-oxobutanoic acid, belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. L-2-Amino-3-oxobutanoic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. L-2-Amino-3-oxobutanoic acid exists in all living species, ranging from bacteria to humans. L-2-Amino-3-oxobutanoic acid can be biosynthesized from acetyl-CoA and glycine through the action of the enzyme L-L-l-2-amino-3-oxobutanoic acid coenzyme A ligase, mitochondrial. In cattle, L-L-l-2-amino-3-oxobutanoic acid is involved in the metabolic pathway called the glycine and serine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2-Amino-3-oxobutanoic acid | ChEBI | | 2-AMINO-3-ketobutyrIC ACID | ChEBI | | L-2-Amino-3-oxobutanoate | ChEBI | | L-2-Amino-acetoacetate | ChEBI | | (S)-2-Amino-3-oxobutanoate | Generator | | 2-AMINO-3-ketobutyrate | Generator | | L-2-Amino-acetoacetic acid | Generator | | (2S)-2-amino-3-Oxobutanoate | HMDB | | (2S)-2-amino-3-Oxobutanoic acid | HMDB | | 2-amino-3-Oxobutanoate | HMDB | | 2-amino-3-Oxobutanoic acid | HMDB | | L-2-amino Acetic acid | HMDB |
|
|---|
| Chemical Formula | C4H7NO3 |
|---|
| Average Molecular Weight | 117.1033 |
|---|
| Monoisotopic Molecular Weight | 117.042593095 |
|---|
| IUPAC Name | (2S)-2-amino-3-oxobutanoic acid |
|---|
| Traditional Name | 2-amino-3-ketobutyric acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CC(=O)[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H7NO3/c1-2(6)3(5)4(7)8/h3H,5H2,1H3,(H,7,8)/t3-/m0/s1 |
|---|
| InChI Key | SAUCHDKDCUROAO-VKHMYHEASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Beta-keto acid
- Short-chain keto acid
- Beta-hydroxy ketone
- Keto acid
- 1,3-dicarbonyl compound
- Alpha-aminoketone
- Amino acid
- Ketone
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Primary aliphatic amine
- Primary amine
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-006x-9000000000-22150e9e4fa3d3433203 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-006y-9300000000-b1976cb0fd0e419923c4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gi0-5900000000-1565f4db4a91c43d6697 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fk9-9200000000-1dcade0f25d1eaa7da38 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pb9-9100000000-ef438e90f9a367453dd6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-9800000000-e0bdda855b20afe7598c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9200000000-95fb6afa14c1b9b751e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9000000000-210f3affcabb1713c4e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-9000000000-b0afa4195d3cbff82f87 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9000000000-c542fb4af78140fc2eaa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-5136afe8f7ab28631fe4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01b9-4900000000-cbd7caa68b545994551b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dj-9200000000-45cde5b7b6c52af1dee2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-1e9dea60f3fc6a6303f7 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|