| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:23:26 UTC |
|---|
| Update Date | 2020-04-22 15:17:53 UTC |
|---|
| BMDB ID | BMDB0006565 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Sialyl-Lewis X |
|---|
| Description | Sialyl-Lewis X belongs to the class of organic compounds known as glutamine and derivatives. Glutamine and derivatives are compounds containing glutamine or a derivative thereof resulting from reaction of glutamine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. Based on a literature review very few articles have been published on Sialyl-Lewis X. |
|---|
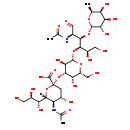
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| SLex | ChEMBL, HMDB | | Sialyl lewisx | ChEMBL, HMDB | | Sialyl lewis-X | ChEMBL, HMDB | | Sialyl lex | ChEMBL, HMDB | | 3'-Sialyl-lewis X | HMDB | | 3'-Sialyl-lewis-X tetrasaccharide | HMDB | | 3'-SLeX | HMDB | | a-NeuNAc-(2>3)-b-D-gal-(1>4)(a-L-fuc-[1>3])-D-glcnac | HMDB | | alpha-NeuNAc-(2>3)-beta-delta-gal-(1>4)(a-L-fuc-[1>3])-delta-glcnac | HMDB | | Sialyl lex tri | HMDB | | SSEA 1 | HMDB, MeSH | | 3 alpha Fucosyl N acetyl lactosamine | MeSH, HMDB | | 3 alpha-Fucosyl-N-acetyl lactosamine | MeSH, HMDB | | Antigen, lewis X | MeSH, HMDB | | Antigens, CD15 | MeSH, HMDB | | CD15 Antigen | MeSH, HMDB | | CD15 Antigens | MeSH, HMDB | | Embryonic antigen-1, stage-specific | MeSH, HMDB | | Galbeta(1-4)fucalpha(1-3)glcnac | MeSH, HMDB | | Hapten X | MeSH, HMDB | | Le(X) antigen | MeSH, HMDB | | Leu m1 antigens | MeSH, HMDB | | Leu-m1 antigens | MeSH, HMDB | | Lewis X antigen | MeSH, HMDB | | Lewis X hapten | MeSH, HMDB | | SSEA 1 determinant | MeSH, HMDB | | SSEA-1 | MeSH, HMDB | | SSEA-1 determinant | MeSH, HMDB | | Stage specific embryonic antigen 1 | MeSH, HMDB | | Stage-specific embryonic antigen-1 | MeSH, HMDB | | X Antigen, lewis | MeSH, HMDB | | X Hapten, lewis | MeSH, HMDB | | (2S,4S,5R,6R)-2-{[(2S,3R,4S,5S,6R)-2-{[(2R,3R,4R,5R)-1,2-dihydroxy-5-[(1-hydroxyethylidene)amino]-6-oxo-4-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}hexan-3-yl]oxy}-3,5-dihydroxy-6-(hydroxymethyl)oxan-4-yl]oxy}-4-hydroxy-5-[(1-hydroxyethylidene)amino]-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylate | Generator, HMDB |
|
|---|
| Chemical Formula | C31H52N2O23 |
|---|
| Average Molecular Weight | 820.7442 |
|---|
| Monoisotopic Molecular Weight | 820.29608598 |
|---|
| IUPAC Name | (2S,4S,5R,6R)-5-acetamido-2-{[(2S,3R,4S,5S,6R)-2-{[(2R,3R,4R,5R)-5-acetamido-1,2-dihydroxy-6-oxo-4-{[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}hexan-3-yl]oxy}-3,5-dihydroxy-6-(hydroxymethyl)oxan-4-yl]oxy}-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylic acid |
|---|
| Traditional Name | sialyl-lewis X |
|---|
| CAS Registry Number | 98603-84-0 |
|---|
| SMILES | [H][C@]1(O[C@@](C[C@H](O)[C@H]1NC(C)=O)(O[C@H]1[C@@H](O)[C@@H](CO)O[C@@H](O[C@H]([C@H](O)CO)[C@H](O[C@@H]2O[C@@H](C)[C@@H](O)[C@@H](O)[C@@H]2O)[C@@H](NC(C)=O)C=O)[C@@H]1O)C(O)=O)[C@H](O)[C@H](O)CO |
|---|
| InChI Identifier | InChI=1S/C31H52N2O23/c1-9-18(43)21(46)22(47)28(51-9)53-24(12(5-34)32-10(2)38)25(15(42)7-36)54-29-23(48)27(20(45)16(8-37)52-29)56-31(30(49)50)4-13(40)17(33-11(3)39)26(55-31)19(44)14(41)6-35/h5,9,12-29,35-37,40-48H,4,6-8H2,1-3H3,(H,32,38)(H,33,39)(H,49,50)/t9-,12-,13-,14+,15+,16+,17+,18+,19+,20-,21+,22-,23+,24+,25+,26+,27-,28-,29-,31-/m0/s1 |
|---|
| InChI Key | LAQPKDLYOBZWBT-NYLDSJSYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamine and derivatives. Glutamine and derivatives are compounds containing glutamine or a derivative thereof resulting from reaction of glutamine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamine or derivatives
- Glutamic acid or derivatives
- Hippuric acid or derivatives
- Pterin
- Alpha-amino acid
- Aminobenzamide
- Aminobenzoic acid or derivatives
- Pteridine
- L-alpha-amino acid
- Benzoic acid or derivatives
- Benzoyl
- Phenylalkylamine
- Aniline or substituted anilines
- Gamma-keto acid
- Aminopyrimidine
- Aralkylamine
- Beta-keto acid
- Amino fatty acid
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Gamma-aminoketone
- Pyrimidine
- Pyrazine
- Benzenoid
- Fatty acyl
- Monocyclic benzene moiety
- Keto acid
- N-acyl-amine
- Carboxylic acid imide, n-substituted
- Heteroaromatic compound
- Carboxylic acid anhydride
- Carboxylic acid imide
- Dicarboximide
- Vinylogous amide
- Ketone
- Amino acid
- Secondary amine
- Carboxylic acid
- Azacycle
- Organoheterocyclic compound
- Organonitrogen compound
- Carbonyl group
- Amine
- Organic oxygen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organic oxide
- Primary aliphatic amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0zfr-0039047780-bd9a483433a6b32b3f21 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-1219050000-55b605a166d86dbfb321 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0hu0-2196012000-f5b9e98bca31bd11ab30 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0kml-3812003940-5ce8467310926be298ca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0pdr-9657230210-501b68e6e411f1adc5e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-8459000000-c06d6848a870f6c1eded | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0le9-7140001890-9a9898e9a80d6b2a0f5d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-053r-3470032930-399f53553692504e461f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-3092000000-571b2edb7a0141c71ab6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fk9-0003201290-ff78595d0f6dd1cd8d54 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0kn9-0394158260-4888247c7630f5b9b88a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01pa-3920000100-e100073c507f9d9a271e | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|