| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:25:57 UTC |
|---|
| Update Date | 2020-05-21 16:28:46 UTC |
|---|
| BMDB ID | BMDB0006779 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Indole-5,6-quinone |
|---|
| Description | Indole-5,6-quinone belongs to the class of organic compounds known as indoles and derivatives. These are organic compounds containing an indole, which is a bicyclic ring system made up of a six-membered benzene ring fused to a five-membered nitrogen-containing pyrrole ring. Indole-5,6-quinone is possibly soluble (in water) and a strong basic compound (based on its pKa). Indole-5,6-quinone exists in all living organisms, ranging from bacteria to humans. |
|---|
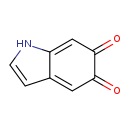
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1H-Indole-5,6-dione | HMDB | | Indole-5,6 quinone | HMDB |
|
|---|
| Chemical Formula | C8H5NO2 |
|---|
| Average Molecular Weight | 147.1308 |
|---|
| Monoisotopic Molecular Weight | 147.032028409 |
|---|
| IUPAC Name | 5,6-dihydro-1H-indole-5,6-dione |
|---|
| Traditional Name | indole-5,6-quinone |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | O=C1C=C2NC=CC2=CC1=O |
|---|
| InChI Identifier | InChI=1S/C8H5NO2/c10-7-3-5-1-2-9-6(5)4-8(7)11/h1-4,9H |
|---|
| InChI Key | IGGVVGHJSQSLFO-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as indoles and derivatives. These are organic compounds containing an indole, which is a bicyclic ring system made up of a six-membered benzene ring fused to a five-membered nitrogen-containing pyrrole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Indoles and derivatives |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Indoles and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Indole or derivatives
- Pyrrole
- Vinylogous amide
- Heteroaromatic compound
- Ketone
- Cyclic ketone
- Azacycle
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-066v-6900000000-1dca4f608ca530016412 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-0cf3a823a731cd4e7a64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dj-0900000000-81eabafffbd73c3732d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-9400000000-a1268508f67a658cecf5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-efe9cb1e2b58b04208a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0900000000-37f54dda461b2c0102c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014j-0900000000-775f13193ba5fc2154eb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-9731758a758f9f924b40 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006t-1900000000-8e368a3b1e151aabfbfa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0fxx-9700000000-616921cda808497b0d2c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-7c00c3e5508255ca8248 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0900000000-a13b820d1ef5510928d7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kb-0900000000-e18791078884a5a972e8 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Qian L, Zhao A, Zhang Y, Chen T, Zeisel SH, Jia W, Cai W: Metabolomic Approaches to Explore Chemical Diversity of Human Breast-Milk, Formula Milk and Bovine Milk. Int J Mol Sci. 2016 Dec 17;17(12). pii: ijms17122128. doi: 10.3390/ijms17122128. [PubMed:27999311 ]
|
|---|