| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:26:17 UTC |
|---|
| Update Date | 2020-05-21 16:28:55 UTC |
|---|
| BMDB ID | BMDB0006809 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Nicotinate D-ribonucleoside |
|---|
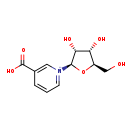
| Description | Nicotinate D-ribonucleoside, also known as nicotinate d-ribonucleoside or beta-Nicotinate d-ribonucleoside, belongs to the class of organic compounds known as glycosylamines. Glycosylamines are compounds consisting of an amine with a beta-N-glycosidic bond to a carbohydrate, thus forming a cyclic hemiaminal ether bond (alpha-amino ether). Nicotinate D-ribonucleoside is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). Nicotinate D-ribonucleoside exists in all living species, ranging from bacteria to humans. Nicotinate D-ribonucleoside participates in a number of enzymatic reactions, within cattle. In particular, Nicotinate D-ribonucleoside can be converted into nicotinic acid mononucleotide through the action of the enzyme nicotinamide riboside kinase 1. In addition, Nicotinate D-ribonucleoside can be biosynthesized from nicotinic acid mononucleotide; which is catalyzed by the enzyme cytosolic purine 5'-nucleotidase. In cattle, nicotinate D-ribonucleoside is involved in the metabolic pathway called the nicotinate and nicotinamide metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Nicotinic acid riboside | ChEBI | | beta-D-Ribosylnicotinate | Kegg | | Nicotinate riboside | Generator | | b-D-Ribosylnicotinate | Generator | | b-D-Ribosylnicotinic acid | Generator | | beta-D-Ribosylnicotinic acid | Generator | | Β-D-ribosylnicotinate | Generator | | Β-D-ribosylnicotinic acid | Generator | | Nicotinic acid D-ribonucleoside | Generator | | D-Ribosylnicotinate | HMDB | | Nicotinic acid ribose | HMDB | | Ribosylnicotinate | HMDB | | Nicotinate ribonucleoside | HMDB | | Nicotinate ribose | HMDB | | Nicotinic acid ribonucleoside | HMDB | | 3-Carboxy-1-beta-D-ribofuranosylpyridinium | HMDB | | 3-Carboxy-1-β-D-ribofuranosylpyridinium | HMDB | | Nicotinic riboside | HMDB |
|
|---|
| Chemical Formula | C11H14NO6 |
|---|
| Average Molecular Weight | 256.232 |
|---|
| Monoisotopic Molecular Weight | 256.082112185 |
|---|
| IUPAC Name | 3-carboxy-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1lambda5-pyridin-1-ylium |
|---|
| Traditional Name | 3-carboxy-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1lambda5-pyridin-1-ylium |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)[N+]1=CC(=CC=C1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C11H13NO6/c13-5-7-8(14)9(15)10(18-7)12-3-1-2-6(4-12)11(16)17/h1-4,7-10,13-15H,5H2/p+1/t7-,8-,9-,10-/m1/s1 |
|---|
| InChI Key | PUEDDPCUCPRQNY-ZYUZMQFOSA-O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycosylamines. Glycosylamines are compounds consisting of an amine with a beta-N-glycosidic bond to a carbohydrate, thus forming a cyclic hemiaminal ether bond (alpha-amino ether). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glycosylamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-glycosyl compound
- Pentose monosaccharide
- Pyridine carboxylic acid
- Pyridine carboxylic acid or derivatives
- Monosaccharide
- Pyridine
- Pyridinium
- Vinylogous amide
- Heteroaromatic compound
- Tetrahydrofuran
- Secondary alcohol
- Organoheterocyclic compound
- Carboxylic acid derivative
- Oxacycle
- Azacycle
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organonitrogen compound
- Alcohol
- Primary alcohol
- Organic nitrogen compound
- Organic cation
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-009f-9640000000-9ecd90015c00728b3b07 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0fa9-9442740000-0573803f81575b22f5c8 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0190000000-c2d0a64c0eb3d4ae2bfa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0490000000-bb44dd196c6e9a647df7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0zgu-9600000000-d17b549da8f5e8b2dd2b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0090000000-384ec325284b36db888b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-002b-2190000000-5ab972cf9d81893b8388 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9510000000-8e2f234fa7933273a714 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06e9-1950000000-ae12a3b253bcd6d48ccc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9510000000-cb2cf76989881501793c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9100000000-9d7edc1ec5f0c17e5711 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|