| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:26:29 UTC |
|---|
| Update Date | 2020-05-21 16:28:32 UTC |
|---|
| BMDB ID | BMDB0006826 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Tetrahydrofolyl-[Glu](n) |
|---|
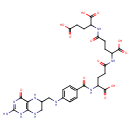
| Description | Tetrahydrofolyl-[glu](N), also known as THF-L-glutamic acid or THF-polyglutamate, belongs to the class of organic compounds known as tetrahydrofolic acids and derivatives. These are heterocyclic compounds based on the 5,6,7,8-tetrahydropteroic acid skeleton conjugated with at least one L-glutamic acid unit (or a derivative thereof) . Tetrahydrofolyl-[glu](N) is possibly soluble (in water) and a strong basic compound (based on its pKa). Tetrahydrofolyl-[glu](N) participates in a number of enzymatic reactions, within cattle. In particular, Tetrahydrofolyl-[glu](N) can be biosynthesized from tetrahydrofolyl-[glu](2) and L-glutamic acid; which is catalyzed by the enzyme folylpolyglutamate synthase. In addition, Tetrahydrofolyl-[glu](N) can be converted into tetrahydrofolyl-[glu](2) and L-glutamic acid; which is catalyzed by the enzyme Gamma-glutamyl hydrolase. In cattle, tetrahydrofolyl-[glu](N) is involved in the metabolic pathway called the folate metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Tetrahydrofolyl-[glu](n+1) | HMDB | | Tetrahydropteroyl-[gamma-glu]N | HMDB | | Tetrahydropteroyl-[gamma-glu]n+1 | HMDB | | THF-L-Glutamic acid | HMDB | | THF-Polyglutamate | HMDB |
|
|---|
| Chemical Formula | C29H37N9O12 |
|---|
| Average Molecular Weight | 703.6572 |
|---|
| Monoisotopic Molecular Weight | 703.256167693 |
|---|
| IUPAC Name | 2-(4-{4-[(4-{[(2-amino-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl)methyl]amino}phenyl)formamido]-4-carboxybutanamido}-4-carboxybutanamido)pentanedioic acid |
|---|
| Traditional Name | 2-(4-{4-[(4-{[(2-amino-4-oxo-5,6,7,8-tetrahydro-1H-pteridin-6-yl)methyl]amino}phenyl)formamido]-4-carboxybutanamido}-4-carboxybutanamido)pentanedioic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C=C3)C(=O)NC(CCC(=O)NC(CCC(=O)NC(CCC(O)=O)C(O)=O)C(O)=O)C(O)=O)N2)N1 |
|---|
| InChI Identifier | InChI=1S/C29H37N9O12/c30-29-37-23-22(25(44)38-29)33-15(12-32-23)11-31-14-3-1-13(2-4-14)24(43)36-18(28(49)50)6-9-20(40)34-16(26(45)46)5-8-19(39)35-17(27(47)48)7-10-21(41)42/h1-4,15-18,31,33H,5-12H2,(H,34,40)(H,35,39)(H,36,43)(H,41,42)(H,45,46)(H,47,48)(H,49,50)(H4,30,32,37,38,44) |
|---|
| InChI Key | RXWVHRYZTWZATH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tetrahydrofolic acids and derivatives. These are heterocyclic compounds based on the 5,6,7,8-tetrahydropteroic acid skeleton conjugated with at least one L-glutamic acid unit (or a derivative thereof) . |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Tetrahydrofolic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tetrahydrofolic acid or derivatives
- Alpha-oligopeptide
- Gamma-glutamyl alpha-amino acid
- Glutamic acid or derivatives
- Glutamine or derivatives
- Tetracarboxylic acid or derivatives
- Hippuric acid or derivatives
- Hippuric acid
- N-acyl-alpha amino acid or derivatives
- N-acyl-alpha-amino acid
- Aminobenzamide
- Aminobenzoic acid or derivatives
- Alpha-amino acid or derivatives
- Benzoic acid or derivatives
- Benzamide
- Phenylalkylamine
- Aniline or substituted anilines
- Benzoyl
- Aminopyrimidine
- Pyrimidone
- Secondary aliphatic/aromatic amine
- N-acyl-amine
- Pyrimidine
- Fatty acyl
- Monocyclic benzene moiety
- Fatty amide
- Benzenoid
- Vinylogous amide
- Heteroaromatic compound
- Amino acid or derivatives
- Amino acid
- Secondary carboxylic acid amide
- Carboxamide group
- Carboxylic acid
- Carboxylic acid derivative
- Azacycle
- Secondary amine
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Amine
- Organic oxide
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-0330229000-26f7358d09d8be8dd6bc | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-053i-0330009000-2343f1529d5558b72f37 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0960106000-948a11df3216bc17a492 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0089-0930102000-c63ca5bc90715eab8c7b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0kal-0000009200-7ef523ed0c34682f53c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06rx-0110119000-94abb8fbe6ce9db951ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-054p-6941203000-2220ce26cc38dbfa7133 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0001009700-73c689fed96633d0e3c4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ufr-0921105100-6c0b5caba5aa9d72304b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ufr-1910000000-3587158aa91827b1a1f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f72-0190125500-66fcaf7dd19aada1c2a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-055e-0890461000-4f80e46b7c8bdfa76947 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01q9-1960030000-fb370cb9f66800f069e9 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|