| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:26:58 UTC |
|---|
| Update Date | 2020-05-21 16:28:45 UTC |
|---|
| BMDB ID | BMDB0006868 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | S-(2-Methylpropionyl)-dihydrolipoamide-E |
|---|
| Description | S-(2-Methylpropionyl)-dihydrolipoamide-e belongs to the class of organic compounds known as fatty amides. These are carboxylic acid amide derivatives of fatty acids, that are formed from a fatty acid and an amine. Thus, S-(2-methylpropionyl)-dihydrolipoamide-e-e is considered to be a fatty amide lipid molecule. S-(2-Methylpropionyl)-dihydrolipoamide-e is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. S-(2-Methylpropionyl)-dihydrolipoamide-e participates in a number of enzymatic reactions, within cattle. In particular, S-(2-Methylpropionyl)-dihydrolipoamide-e can be biosynthesized from 2-methyl-1-hydroxypropyl-THPP through the action of the enzymes 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial and 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial. In addition, S-(2-Methylpropionyl)-dihydrolipoamide-e can be biosynthesized from lipoamide through the action of the enzymes 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial and 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial. In cattle, S-(2-methylpropionyl)-dihydrolipoamide-e-e is involved in the metabolic pathway called the valine, leucine, and isoleucine degradation pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| S-(2-Methylpropanoyl)-dihydrolipoamide | ChEBI | | S-(2-Methylpropionyl)-dihydrolipoamide | ChEBI | | S-(2-Methylpropanoyl)-dihydrolipoamide-e | HMDB | | [Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase]S-(2-methylpropanoyl)dihydrolipoyllysine | HMDB |
|
|---|
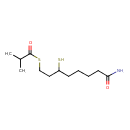
| Chemical Formula | C12H23NO2S2 |
|---|
| Average Molecular Weight | 277.447 |
|---|
| Monoisotopic Molecular Weight | 277.117020365 |
|---|
| IUPAC Name | 8-[(2-methylpropanoyl)sulfanyl]-6-sulfanyloctanamide |
|---|
| Traditional Name | 8-[(2-methylpropanoyl)sulfanyl]-6-sulfanyloctanamide |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CC(C)C(=O)SCCC(S)CCCCC(N)=O |
|---|
| InChI Identifier | InChI=1S/C12H23NO2S2/c1-9(2)12(15)17-8-7-10(16)5-3-4-6-11(13)14/h9-10,16H,3-8H2,1-2H3,(H2,13,14) |
|---|
| InChI Key | UEFURMXXHJCLJP-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty amides. These are carboxylic acid amide derivatives of fatty acids, that are formed from a fatty acid and an amine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty amides |
|---|
| Direct Parent | Fatty amides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty amide
- Carboxamide group
- Primary carboxylic acid amide
- Thiocarboxylic acid ester
- Carbothioic s-ester
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Alkylthiol
- Carboxylic acid derivative
- Carbonyl group
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-006x-9430000000-898ad5ac7f8d34382fce | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0200-1590000000-256cf1b407c15039646a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05fr-7960000000-68678b3f077aeb471401 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0k96-9600000000-b142c6b121e61d161a14 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-062c-3590000000-dd8c6dd74f1ee11d43e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0axu-6790000000-b1e8d0173e84691b6bbb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9000000000-ec64c5922fd146f463b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0190000000-7ce7db6d296a0c9ccd1c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-044l-3950000000-408139b7c147c5ffb59b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-007d-9400000000-69ca2ae5bb862cef6baf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-8ed120c1f1da33069a71 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zmi-2940000000-f72c23450f1a5b08d40c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-daf484d53de0437afb9f | View in MoNA |
|---|
|
|---|