| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:31:08 UTC |
|---|
| Update Date | 2020-06-04 19:52:09 UTC |
|---|
| BMDB ID | BMDB0007128 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | DG(16:1(9Z)/16:1(9Z)/0:0) |
|---|
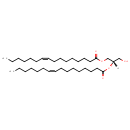
| Description | DG(16:1(9Z)/16:1(9Z)/0:0), also known as diacylglycerol or DAG(16:1/16:1), belongs to the class of organic compounds known as 1,2-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 2. Thus, DG(16:1(9Z)/16:1(9Z)/0:0) is considered to be a diradylglycerol lipid molecule. DG(16:1(9Z)/16:1(9Z)/0:0) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. DG(16:1(9Z)/16:1(9Z)/0:0) exists in all living organisms, ranging from bacteria to humans. DG(16:1(9Z)/16:1(9Z)/0:0) participates in a number of enzymatic reactions, within cattle. In particular, CDP-Ethanolamine and DG(16:1(9Z)/16:1(9Z)/0:0) can be converted into cytidine monophosphate and PE(16:1(9Z)/16:1(9Z)); which is catalyzed by the enzyme choline/ethanolaminephosphotransferase. In addition, CDP-Choline and DG(16:1(9Z)/16:1(9Z)/0:0) can be converted into cytidine monophosphate and PC(16:1(9Z)/16:1(9Z)) through the action of the enzyme choline/ethanolaminephosphotransferase. In cattle, DG(16:1(9Z)/16:1(9Z)/0:0) is involved in the metabolic pathway called phosphatidylcholine biosynthesis PC(16:1(9Z)/16:1(9Z)) pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2-Di-(9Z-hexadecenoyl)-sn-glycerol | ChEBI | | DAG(16:1/16:1) | ChEBI | | DAG(16:1N7/16:1N7) | ChEBI | | DAG(32:2) | ChEBI | | DG(16:1/16:1/0:0) | ChEBI | | DG(16:1W7/16:1W7) | ChEBI | | DG(32:2) | ChEBI | | 1,2-dipalmitoleoyl-rac-glycerol | Lipid Annotator, HMDB | | DG(16:1(9Z)/16:1(9Z)/0:0) | Lipid Annotator | | Diglyceride | Lipid Annotator, HMDB | | DG(16:1/16:1) | Lipid Annotator, HMDB | | Diacylglycerol | Lipid Annotator, HMDB | | Diacylglycerol(16:1/16:1) | Lipid Annotator, HMDB | | 1,2-di(9Z-hexadecenoyl)-rac-glycerol | Lipid Annotator, HMDB | | Diacylglycerol(32:2) | Lipid Annotator, HMDB |
|
|---|
| Chemical Formula | C35H64O5 |
|---|
| Average Molecular Weight | 564.8797 |
|---|
| Monoisotopic Molecular Weight | 564.475375158 |
|---|
| IUPAC Name | (2S)-1-[(9Z)-hexadec-9-enoyloxy]-3-hydroxypropan-2-yl (9Z)-hexadec-9-enoate |
|---|
| Traditional Name | diacylglycerol |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H][C@](CO)(COC(=O)CCCCCCC\C=C/CCCCCC)OC(=O)CCCCCCC\C=C/CCCCCC |
|---|
| InChI Identifier | InChI=1S/C35H64O5/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-34(37)39-32-33(31-36)40-35(38)30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h13-16,33,36H,3-12,17-32H2,1-2H3/b15-13-,16-14-/t33-/m0/s1 |
|---|
| InChI Key | HSQHRRHRYJNSOC-DWCRPSDDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Diradylglycerols |
|---|
| Direct Parent | 1,2-diacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-acyl-sn-glycerol
- Fatty acid ester
- Fatty acyl
- Dicarboxylic acid or derivatives
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-008i-4449464000-50b6a5cf704160fba39d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("DG(16:1(9Z)/16:1(9Z)/0:0),1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000090000-51d305f10f90afe4eb07 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dj-0009090000-367853de731f7e261d22 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03e9-0009090000-736308809e6d3cc751f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0043090000-9be462094b27393e9285 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0w2i-2092000000-a9d9a5b97c3b76b05af0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udr-1290000000-987ad4e1a1321117b546 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-2244390000-c5ba870493b35853231d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-7294120000-5fb9d214e1f99e650a8f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06rj-9363000000-500eddc99602756cec87 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000090000-279f8e7a1315590373dc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dj-0008090000-54ee6ee0a058250d5412 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03e9-0008090000-cd2f6f1c784cfcb067d1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000090000-ec1b73afc18d0387d07f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000090000-ec1b73afc18d0387d07f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-0019010000-2a8ef0425a75fbd6ee6c | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|