| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:31:19 UTC |
|---|
| Update Date | 2020-05-21 16:27:44 UTC |
|---|
| BMDB ID | BMDB0007137 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | DG(16:1(9Z)/20:1(11Z)/0:0) |
|---|
| Description | DG(16:1(9Z)/20:1(11Z)/0:0), also known as diacylglycerol(36:2) or DAG(16:1/20:1), belongs to the class of organic compounds known as 1,2-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 2. Thus, DG(16:1(9Z)/20:1(11Z)/0:0) is considered to be a diradylglycerol lipid molecule. DG(16:1(9Z)/20:1(11Z)/0:0) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
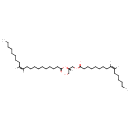
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| DG(36:2) | HMDB | | Diacylglycerol(36:2) | HMDB | | DAG(16:1/20:1) | HMDB | | DAG(36:2) | HMDB | | DG(16:1/20:1) | HMDB | | Diglyceride | HMDB | | Diacylglycerol | HMDB | | Diacylglycerol(16:1/20:1) | HMDB | | 1-(9Z-Hexadecenoyl)-2-(11-eicosenoyl)-sn-glycerol | HMDB | | 1-Palmitoleoyl-2-eicosenoyl-sn-glycerol | HMDB | | DG(16:1(9Z)/20:1(11Z)/0:0) | Lipid Annotator |
|
|---|
| Chemical Formula | C39H72O5 |
|---|
| Average Molecular Weight | 620.986 |
|---|
| Monoisotopic Molecular Weight | 620.537975414 |
|---|
| IUPAC Name | (2S)-1-[(9Z)-hexadec-9-enoyloxy]-3-hydroxypropan-2-yl (11Z)-icos-11-enoate |
|---|
| Traditional Name | diacylglycerol |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H]\C(CCCCCC)=C(/[H])CCCCCCCC(=O)OC[C@]([H])(CO)OC(=O)CCCCCCCCC\C([H])=C(\[H])CCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C39H72O5/c1-3-5-7-9-11-13-15-17-18-19-20-22-24-26-28-30-32-34-39(42)44-37(35-40)36-43-38(41)33-31-29-27-25-23-21-16-14-12-10-8-6-4-2/h14,16-18,37,40H,3-13,15,19-36H2,1-2H3/b16-14-,18-17-/t37-/m0/s1 |
|---|
| InChI Key | FBCDXXQBGHZSLI-JSTCSLLMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Diradylglycerols |
|---|
| Direct Parent | 1,2-diacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-acyl-sn-glycerol
- Fatty acid ester
- Fatty acyl
- Dicarboxylic acid or derivatives
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00ac-4492357000-3d22ff51f4a801344609 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("DG(16:1(9Z)/20:1(11Z)/0:0),1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000009000-9185b686832caa4f55c4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0jor-0009004000-ad6e796024c8b0bc3066 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-04i9-0009004000-f278b3d85a1fbc5c5a31 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0303-1049005000-dc4297d5e07505f35bb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kf-3398001000-488d1184aac3d820ebcd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000x-9411000000-ecc5cccf567c4778aa8c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000009000-203520884a3c62e5734f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0jor-0009004000-de60fb455f6fdefb6396 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-04i9-0009004000-e43e8d6dcf4266929784 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0aor-1009004000-37ed1ca21909fa8f5cb6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-4097001000-5f1be4ba5e2af72bec87 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-4891000000-faf574895230ab908735 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000009000-84cf426a5bd9b351e99a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-0000009000-84cf426a5bd9b351e99a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001r-0009000000-c808dc8955bacb1234db | View in MoNA |
|---|
|
|---|
| Pathways | |
|---|