| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:32:34 UTC |
|---|
| Update Date | 2020-05-21 16:27:49 UTC |
|---|
| BMDB ID | BMDB0007199 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) |
|---|
| Description | DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0), also known as DG(18:1/20:4) or dg(18:1(11z)/20:4(5z,8z,11z,14z)/0:0), belongs to the class of organic compounds known as 1,2-dg(18:1(11z)/20:4(5z,8z,11z,14z)/0:0)s. These are dg(18:1(11z)/20:4(5z,8z,11z,14z)/0:0)s containing a glycerol acylated at positions 1 and 2. DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) participates in a number of enzymatic reactions, within cattle. In particular, DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) can be biosynthesized from PA(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)) through its interaction with the enzyme phosphatidate phosphatase. Furthermore, DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) and docosanoyl-CoA can be converted into TG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/22:0); which is catalyzed by the enzyme dg(18:1(11z)/20:4(5z,8z,11z,14z)/0:0) O-acyltransferase. Furthermore, DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) can be biosynthesized from PA(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)); which is catalyzed by the enzyme phosphatidate phosphatase. Finally, DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) and tetracosanoyl-CoA can be converted into TG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/24:0); which is mediated by the enzyme dg(18:1(11z)/20:4(5z,8z,11z,14z)/0:0) O-acyltransferase. In cattle, DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) is involved in a couple of metabolic pathways, which include de novo triacylglycerol biosynthesis TG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/22:0) pathway and de novo triacylglycerol biosynthesis TG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/24:0) pathway. |
|---|
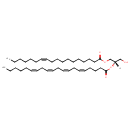
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Vaccenoyl-2-arachidonoyl-sn-glycerol | HMDB | | DG(18:1/20:4) | HMDB | | DG(38:5) | HMDB | | DAG(38:5) | HMDB | | DAG(18:1/20:4) | HMDB | | Diacylglycerol(38:5) | HMDB | | Diacylglycerol | HMDB | | Diacylglycerol(18:1/20:4) | HMDB | | 1-(11Z-Octadecenoyl)-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycerol | HMDB | | Diglyceride | HMDB | | DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0) | Lipid Annotator |
|
|---|
| Chemical Formula | C41H70O5 |
|---|
| Average Molecular Weight | 642.9915 |
|---|
| Monoisotopic Molecular Weight | 642.52232535 |
|---|
| IUPAC Name | (2S)-1-hydroxy-3-[(11Z)-octadec-11-enoyloxy]propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate |
|---|
| Traditional Name | diacylglycerol |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | [H][C@](CO)(COC(=O)CCCCCCCCC\C=C/CCCCCC)OC(=O)CCC\C=C/C\C=C/C\C=C/C\C=C/CCCCC |
|---|
| InChI Identifier | InChI=1S/C41H70O5/c1-3-5-7-9-11-13-15-17-19-20-22-24-26-28-30-32-34-36-41(44)46-39(37-42)38-45-40(43)35-33-31-29-27-25-23-21-18-16-14-12-10-8-6-4-2/h11,13-14,16-17,19,22,24,28,30,39,42H,3-10,12,15,18,20-21,23,25-27,29,31-38H2,1-2H3/b13-11-,16-14-,19-17-,24-22-,30-28-/t39-/m0/s1 |
|---|
| InChI Key | UUDOMEYLXRMHOC-FXHMYPJMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Diradylglycerols |
|---|
| Direct Parent | 1,2-diacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-acyl-sn-glycerol
- Fatty acid ester
- Fatty acyl
- Dicarboxylic acid or derivatives
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000i-2194040000-612ea0469a984d1a373b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("DG(18:1(11Z)/20:4(5Z,8Z,11Z,14Z)/0:0),1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-052u-1098005000-bfd299d8cab8a45a2f1e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-2094021000-6bac55ab5828b6da3494 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kr-1091020000-3a3c60fd11b50a6a3d8b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0gxc-0095003000-94d590bcfe1f40422b20 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01q9-1093000000-d69d921e4338c9597dbf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0lzi-3092000000-eaf23a7205e4115bba52 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0000009000-25d712cfd42c783e22f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0000009000-25d712cfd42c783e22f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03f0-0009000000-f356af4e18adaf86964c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000l-2094036000-4e556e57c31c074170e2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-4294000000-c29fbcdcec3175010d24 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06ri-3397000000-4a187bbf413378fd32ca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-1046009000-8040c9419e182f5c1181 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01x0-4095001000-3b12c457a5679af53209 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0lz9-4192000000-fcdcee82f000c22a987c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000009000-c42d84986800260de203 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03g0-0009004000-b3797e24691c61ae4c8f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dr-0009004000-738dab90f979fa857ce9 | View in MoNA |
|---|
|
|---|
| Pathways | |
|---|