| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 17:14:12 UTC |
|---|
| Update Date | 2020-05-21 16:28:20 UTC |
|---|
| BMDB ID | BMDB0008123 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
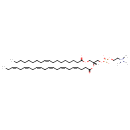
| Common Name | PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
|---|
| Description | PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)), also known as PC(18:1/22:6) or 1-oleoyl-2-docosahexaenoyl-GPC, belongs to the class of organic compounds known as phosphatidylcholines. These are glycerophosphocholines in which the two free -OH are attached to one fatty acid each through an ester linkage. Thus, PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) is considered to be a glycerophosphocholine lipid molecule. PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) participates in a number of enzymatic reactions, within cattle. In particular, S-Adenosylhomocysteine and PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) can be biosynthesized from S-adenosylmethionine and pe-nme2(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)); which is catalyzed by the enzyme phosphatidylethanolamine N-methyltransferase. Furthermore, Cytidine monophosphate and PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) can be biosynthesized from CDP-choline and DG(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)/0:0); which is catalyzed by the enzyme choline/ethanolaminephosphotransferase. Finally, PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) and L-serine can be converted into choline and PS(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)); which is mediated by the enzyme phosphatidylserine synthase. In cattle, PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) is involved in a couple of metabolic pathways, which include phosphatidylcholine biosynthesis PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) pathway and phosphatidylethanolamine biosynthesis pe(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-(9Z-Octadecenoyl)-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phosphocholine | ChEBI | | 1-Oleoyl-2-docosahexaenoyl-GPC | ChEBI | | 1-Oleoyl-2-docosahexaenoyl-GPC (18:1/22:6) | ChEBI | | 1-Oleoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine | ChEBI | | GPC(18:1/22:6) | ChEBI | | GPCho(18:1/22:6) | ChEBI | | GPCho(18:1n9/22:6n3) | ChEBI | | GPCho(18:1W9/22:6W3) | ChEBI | | PC(18:1/22:6) | ChEBI | | PC(18:1n9/22:6n3) | ChEBI | | PC(18:1W9/22:6W3) | ChEBI | | Phosphatidylcholine(18:1/22:6) | ChEBI | | Phosphatidylcholine(18:1n9/22:6n3) | ChEBI | | Phosphatidylcholine(18:1W9/22:6W3) | ChEBI | | 1-Oleoyl-2-docosahexaenoylglycero-3-phosphorylcholine | HMDB | | 1-ODHPC | HMDB | | 1-Oleoyl-2-dha-PC | HMDB | | 1-Oleoyl-2-docosahexaenoyl phosphatidylcholine | HMDB | | GPCho(40:7) | HMDB | | Lecithin | HMDB | | Phosphatidylcholine(40:7) | HMDB | | PC(40:7) | HMDB | | PC(18:1(9Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | Lipid Annotator |
|
|---|
| Chemical Formula | C48H82NO8P |
|---|
| Average Molecular Weight | 832.1403 |
|---|
| Monoisotopic Molecular Weight | 831.577805117 |
|---|
| IUPAC Name | (2-{[(2R)-2-[(4Z,7Z,10Z,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoyloxy]-3-[(9Z)-octadec-9-enoyloxy]propyl phosphonato]oxy}ethyl)trimethylazanium |
|---|
| Traditional Name | lecithin |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CCCCCCCC\C=C/CCCCCCCC(=O)OC[C@]([H])(COP([O-])(=O)OCC[N+](C)(C)C)OC(=O)CC\C=C/C\C=C/C\C=C/C\C=C/C\C=C/C\C=C/CC |
|---|
| InChI Identifier | InChI=1S/C48H82NO8P/c1-6-8-10-12-14-16-18-20-22-23-24-25-27-29-31-33-35-37-39-41-48(51)57-46(45-56-58(52,53)55-43-42-49(3,4)5)44-54-47(50)40-38-36-34-32-30-28-26-21-19-17-15-13-11-9-7-2/h8,10,14,16,20-22,24-26,29,31,35,37,46H,6-7,9,11-13,15,17-19,23,27-28,30,32-34,36,38-45H2,1-5H3/b10-8-,16-14-,22-20-,25-24-,26-21-,31-29-,37-35-/t46-/m1/s1 |
|---|
| InChI Key | FPEVDFOXMCHLKL-KFDYCXSYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phosphatidylcholines. These are glycerophosphocholines in which the two free -OH are attached to one fatty acid each through an ester linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphocholines |
|---|
| Direct Parent | Phosphatidylcholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Diacylglycero-3-phosphocholine
- Phosphocholine
- Fatty acid ester
- Dialkyl phosphate
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Quaternary ammonium salt
- Tetraalkylammonium salt
- Carboxylic acid ester
- Carboxylic acid derivative
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Amine
- Organic salt
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Intracellular membrane
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000000090-79f227f2d9029f28e7e8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0600000090-924f091d218719cd1b75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f89-1900041120-9baee8374a0c3e7329f1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0093000030-ec8656723a30a525c583 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01q9-0091000100-5783b610f5d7b4fd7e30 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06w9-5095000000-a12c963b60b3d03fa275 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0000000090-9ba117d1c0de1c2e67a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0600000090-d0b4cd4d5b1d3c3b81bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ff0-1900041120-ef5e60e7b652c633f0cc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000000090-b0b8959ba582c76807b7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0011000090-b8190d8ba7ea3bb4e466 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01ai-0099000090-8374d15f57f1ded56758 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000000090-620340c637035e66e687 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000000190-3bb76294110b3cbe4d52 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dk-0900179110-bdfdcea3af515e6884d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000000090-9663725765fd23c75bff | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0044004490-1002bc162cfeff2d6df3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-003r-3192000000-0505b81ab7820b9f17d4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000000090-28e9f69759c65edf3511 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0000000190-f4871623ad0b081e303c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-0200498220-6d6cbbe2f4f218dac20a | View in MoNA |
|---|
|
|---|