| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:13:29 UTC |
|---|
| Update Date | 2020-04-22 15:42:53 UTC |
|---|
| BMDB ID | BMDB0011140 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Hypusine |
|---|
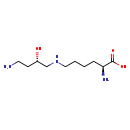
| Description | Hypusine belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. Thus, hypusine is considered to be a fatty acid. Based on a literature review a significant number of articles have been published on Hypusine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N(6)-(4-amino-2-Hydroxybutyl)-L-lysine | HMDB, MeSH | | (R)-N6-(4-Amino-2-hydroxybutyl)-L-lysine | MeSH | | (+)-Hypusine | MeSH |
|
|---|
| Chemical Formula | C10H23N3O3 |
|---|
| Average Molecular Weight | 233.3079 |
|---|
| Monoisotopic Molecular Weight | 233.173941617 |
|---|
| IUPAC Name | (2S)-2-amino-6-{[(2S)-4-amino-2-hydroxybutyl]amino}hexanoic acid |
|---|
| Traditional Name | hypusine |
|---|
| CAS Registry Number | 34994-11-1 |
|---|
| SMILES | NCC[C@H](O)CNCCCC[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C10H23N3O3/c11-5-4-8(14)7-13-6-2-1-3-9(12)10(15)16/h8-9,13-14H,1-7,11-12H2,(H,15,16)/t8-,9-/m0/s1 |
|---|
| InChI Key | BZUIJMCJNWUGKQ-IUCAKERBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Medium-chain fatty acid
- Amino fatty acid
- Hydroxy fatty acid
- Fatty acyl
- Fatty acid
- 1,3-aminoalcohol
- 1,2-aminoalcohol
- Amino acid
- Secondary alcohol
- Carboxylic acid
- Secondary aliphatic amine
- Secondary amine
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Primary aliphatic amine
- Primary amine
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Amine
- Alcohol
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-008c-9300000000-108237316347ea1c9582 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-01x9-9552000000-2e9638e4118872cb5b4e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ba-0970000000-3e60b13fcacb2b767aae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-2910000000-69aafaa99f3bf6faa454 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00e9-9800000000-e00776069e6355a4b997 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0190000000-473d482012482bd6fd27 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03l1-3970000000-cf5864dd6c4bc8e73f8f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fmj-9400000000-51f5a1a2e2551422dae7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-fa9adcf11b63c0cf57e6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01q9-0490000000-f856c8b4b30e39b22442 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-9200000000-be64e780b84a9505cae9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-3390000000-ca6a7e923dc0bfd7383c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9820000000-29d8c010fc356f07f9a4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0089-9400000000-3400136b40459758a26d | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|