| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:21:40 UTC |
|---|
| Update Date | 2020-04-22 15:45:35 UTC |
|---|
| BMDB ID | BMDB0011612 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 6-Hydroxy flavin adenine dinucleotide |
|---|
| Description | 6-Hydroxy flavin adenine dinucleotide, also known as 6-HO-fad, belongs to the class of organic compounds known as flavin nucleotides. These are nucleotides containing a flavin moiety. Flavin is a compound that contains the tricyclic isoalloxazine ring system, which bears 2 oxo groups at the 2- and 4-positions. Based on a literature review a significant number of articles have been published on 6-Hydroxy flavin adenine dinucleotide. |
|---|
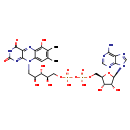
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6-HO-FAD | ChEBI | | 6-HYDROXY-flavin-adenine dinucleotide | ChEBI | | 6-Hydroxyflavin-adenine dinucleotide | ChEBI | | 6-Hydroxyflavine-adenine dinucleotide | ChEBI | | 6-Hydroxyriboflavin 5'-(trihydrogen diphosphate) 5'->5'-ester with adenosine | ChEBI | | 6-OH-Fad | ChEBI | | [(2R,3S,4R,5R)-5-(6-Amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-(6-hydroxy-7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)pentyl dihydrogen diphosphate | ChEBI | | Adenosine 5'-(trihydrogen diphosphate) 5'->5'-ester with 6-hydroxyriboflavin | ChEBI | | 6-Hydroxyriboflavin 5'-(trihydrogen diphosphoric acid) 5'->5'-ester with adenosine | Generator | | [(2R,3S,4R,5R)-5-(6-Amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-(6-hydroxy-7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)pentyl dihydrogen diphosphoric acid | Generator | | Adenosine 5'-(trihydrogen diphosphoric acid) 5'->5'-ester with 6-hydroxyriboflavin | Generator | | 6-Hydroxy-7,8-dimethyl-10-(ribityl-5'-ADP)-isoalloxazine | HMDB | | 6-Hydroxy-fad | HMDB |
|
|---|
| Chemical Formula | C27H33N9O16P2 |
|---|
| Average Molecular Weight | 801.5491 |
|---|
| Monoisotopic Molecular Weight | 801.152049077 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4S)-2,3,4-trihydroxy-5-{6-hydroxy-7,8-dimethyl-2,4-dioxo-2H,3H,4H,10H-benzo[g]pteridin-10-yl}pentyl]oxy})phosphinic acid |
|---|
| Traditional Name | 6-hydroxy-fad |
|---|
| CAS Registry Number | 52301-43-6 |
|---|
| SMILES | CC1=C(C)C(O)=C2N=C3C(=O)NC(=O)N=C3N(C[C@H](O)[C@H](O)[C@H](O)COP(O)(=O)OP(O)(=O)OC[C@H]3O[C@H]([C@H](O)[C@@H]3O)N3C=NC4=C(N)N=CN=C34)C2=C1 |
|---|
| InChI Identifier | InChI=1S/C27H33N9O16P2/c1-9-3-11-15(18(39)10(9)2)32-17-24(33-27(44)34-25(17)43)35(11)4-12(37)19(40)13(38)5-49-53(45,46)52-54(47,48)50-6-14-20(41)21(42)26(51-14)36-8-31-16-22(28)29-7-30-23(16)36/h3,7-8,12-14,19-21,26,37-42H,4-6H2,1-2H3,(H,45,46)(H,47,48)(H2,28,29,30)(H,34,43,44)/t12-,13+,14+,19-,20+,21+,26+/m0/s1 |
|---|
| InChI Key | BJSUUWFQAMLNKU-OKXKTURISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as flavin nucleotides. These are nucleotides containing a flavin moiety. Flavin is a compound that contains the tricyclic isoalloxazine ring system, which bears 2 oxo groups at the 2- and 4-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Flavin nucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Flavin nucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Flavin nucleotide
- (3'->5')-dinucleotide
- (3'->5')-dinucleotide or analogue
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Flavin
- Isoalloxazine
- Pentose-5-phosphate
- Pentose phosphate
- Glycosyl compound
- N-glycosyl compound
- Organic pyrophosphate
- 6-aminopurine
- Diazanaphthalene
- Pentose monosaccharide
- Pteridine
- Quinoxaline
- Monosaccharide phosphate
- Purine
- Imidazopyrimidine
- 1-hydroxy-4-unsubstituted benzenoid
- Monoalkyl phosphate
- Aminopyrimidine
- Pyrimidone
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Imidolactam
- Benzenoid
- Alkyl phosphate
- Phosphoric acid ester
- Pyrimidine
- Pyrazine
- Tetrahydrofuran
- Azole
- Vinylogous amide
- Heteroaromatic compound
- Imidazole
- Secondary alcohol
- Lactam
- Polyol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Organic oxide
- Organic oxygen compound
- Organopnictogen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Primary amine
- Alcohol
- Amine
- Organic nitrogen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0932100210-53f37315ec40b23e0ac8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0931000000-3538e3388fe23d11f6db | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0970000000-25dd99cf0c0180b56d92 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-052f-9382310800-7a79689d89b7001f3285 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-053u-6940100000-f40e5ef5d200d04f3e27 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a59-3910000000-907e26305901d23d35ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000000190-d34a31c9e5de23375bef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uki-0874962580-368434e28e2c047cb143 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0942300000-9d8dd5a3c36f7951d91d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0000000090-760be2b6f7b4852f4f18 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fba-8203110960-65798116bee8b1c6d018 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004j-9423310000-dff8b36d5ccd2e73af04 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|