| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:22:00 UTC |
|---|
| Update Date | 2020-05-21 16:28:50 UTC |
|---|
| BMDB ID | BMDB0011631 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | L-3-Hydroxykynurenine |
|---|
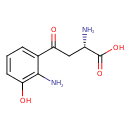
| Description | L-3-Hydroxykynurenine, also known as L-3-hydroxykynurenine, belongs to the class of organic compounds known as alkyl-phenylketones. These are aromatic compounds containing a ketone substituted by one alkyl group, and a phenyl group. L-3-Hydroxykynurenine is possibly soluble (in water) and a very strong basic compound (based on its pKa). L-3-Hydroxykynurenine exists in all living species, ranging from bacteria to humans. L-3-Hydroxykynurenine participates in a number of enzymatic reactions, within cattle. In particular, L-3-Hydroxykynurenine can be biosynthesized from L-kynurenine; which is catalyzed by the enzyme kynurenine 3-monooxygenase. In addition, L-3-Hydroxykynurenine can be converted into 3-hydroxyanthranilic acid and L-alanine through its interaction with the enzyme kynureninase. In cattle, L-3-hydroxykynurenine is involved in the metabolic pathway called the tryptophan metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Amino-4-(2-amino-3-hydroxyphenyl)-4-oxobutanoic acid | ChEBI | | 3-(3-Hydroxyanthraniloyl)-L-alanine | ChEBI | | (2S)-2-Amino-4-(2-amino-3-hydroxyphenyl)-4-oxobutanoate | Generator | | 3-(2-Amino-3-hydroxybenzoyl)-L-alanine | HMDB | | 3-Hydroxy-L-kynurenine | HMDB | | L-3-HK | HMDB |
|
|---|
| Chemical Formula | C10H12N2O4 |
|---|
| Average Molecular Weight | 224.2133 |
|---|
| Monoisotopic Molecular Weight | 224.079706882 |
|---|
| IUPAC Name | (2S)-2-amino-4-(2-amino-3-hydroxyphenyl)-4-oxobutanoic acid |
|---|
| Traditional Name | 3-hydroxy-L-kynurenine |
|---|
| CAS Registry Number | 606-14-4 |
|---|
| SMILES | N[C@@H](CC(=O)C1=CC=CC(O)=C1N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C10H12N2O4/c11-6(10(15)16)4-8(14)5-2-1-3-7(13)9(5)12/h1-3,6,13H,4,11-12H2,(H,15,16)/t6-/m0/s1 |
|---|
| InChI Key | VCKPUUFAIGNJHC-LURJTMIESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alkyl-phenylketones. These are aromatic compounds containing a ketone substituted by one alkyl group, and a phenyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Alkyl-phenylketones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alkyl-phenylketone
- Butyrophenone
- L-alpha-amino acid
- Alpha-amino acid
- Alpha-amino acid or derivatives
- O-aminophenol
- Gamma-keto acid
- Aminophenol
- Aniline or substituted anilines
- Aryl alkyl ketone
- Benzoyl
- Phenol
- 1-hydroxy-2-unsubstituted benzenoid
- 1-hydroxy-4-unsubstituted benzenoid
- Keto acid
- Beta-aminoketone
- Benzenoid
- Monocyclic benzene moiety
- Vinylogous amide
- Amino acid or derivatives
- Amino acid
- Carboxylic acid
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Primary aliphatic amine
- Organonitrogen compound
- Primary amine
- Hydrocarbon derivative
- Organic oxide
- Amine
- Organopnictogen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-057u-7910000000-88ad37433d9fe2009b5e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0596-8379000000-0b0a9595aae2502a55ac | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0960000000-c33d63911a63b0e55059 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0209-2900000000-733debea97f59a15a620 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000l-6900000000-908957a56bc62255065a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-1290000000-49db02e2f97330c6041f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9650000000-45bd0384724255104d6c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ab9-7900000000-e62c0de8b7c63b9e4066 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056r-0890000000-24d8093dfa7cd7471959 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-070i-1900000000-3159845d0fd09fc5476c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9700000000-c823da7e7ec85255fba0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ab9-0690000000-e55cff3d90188bbe1b4f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-1910000000-15ef15dcef2ce9fdfef5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-6900000000-2b99b24490fd7fc7a3f9 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|