| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-10 17:02:09 UTC |
|---|
| Update Date | 2020-04-22 18:56:25 UTC |
|---|
| BMDB ID | BMDB0096158 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Thymidine glycol |
|---|
| Description | Thymidine glycol, also known as glycolthymidine, belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleosides. Pyrimidine 2'-deoxyribonucleosides are compounds consisting of a pyrimidine linked to a ribose which lacks a hydroxyl group at position 2. Based on a literature review very few articles have been published on Thymidine glycol. |
|---|
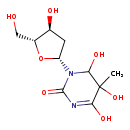
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Thymidine glycol, (5R,6R)-isomer | HMDB | | Thymidine glycol, (cis)-isomer | HMDB | | Thymidine glycol, (trans)-isomer | HMDB | | 5,6-Dihydro-5,6-dihydroxythymidine | HMDB | | Glycolthymidine | HMDB | | Thymidine glycol, (5R*,6S*)-isomer | HMDB | | Thymidine glycol, (5R,6S)-isomer | HMDB | | 5,6-Dihydroxy-5,6-dihydrothymidine | HMDB | | Thymidine glycol, (5S,6R)-isomer | HMDB | | Thymidine glycol, (5S,6S)-isomer | HMDB | | Thymidine glycol | MeSH |
|
|---|
| Chemical Formula | C10H16N2O7 |
|---|
| Average Molecular Weight | 276.2432 |
|---|
| Monoisotopic Molecular Weight | 276.095750876 |
|---|
| IUPAC Name | 4,5,6-trihydroxy-1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,2,5,6-tetrahydropyrimidin-2-one |
|---|
| Traditional Name | 4,5,6-trihydroxy-1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-6H-pyrimidin-2-one |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CC1(O)C(O)N([C@H]2C[C@H](O)[C@@H](CO)O2)C(=O)N=C1O |
|---|
| InChI Identifier | InChI=1S/C10H16N2O7/c1-10(18)7(15)11-9(17)12(8(10)16)6-2-4(14)5(3-13)19-6/h4-6,8,13-14,16,18H,2-3H2,1H3,(H,11,15,17)/t4-,5+,6+,8?,10?/m0/s1 |
|---|
| InChI Key | RKEITGVZZHXKON-SKAWGCAZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleosides. Pyrimidine 2'-deoxyribonucleosides are compounds consisting of a pyrimidine linked to a ribose which lacks a hydroxyl group at position 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleosides |
|---|
| Sub Class | Pyrimidine 2'-deoxyribonucleosides |
|---|
| Direct Parent | Pyrimidine 2'-deoxyribonucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine 2'-deoxyribonucleoside
- Pentose monosaccharide
- Barbiturate
- N-acyl urea
- Pyrimidone
- Ureide
- 1,3-diazinane
- Monosaccharide
- Pyrimidine
- Dicarboximide
- Tetrahydrofuran
- Tertiary alcohol
- Carbonic acid derivative
- Urea
- Secondary alcohol
- Alkanolamine
- Organoheterocyclic compound
- Azacycle
- Carboxylic acid derivative
- Oxacycle
- Organopnictogen compound
- Primary alcohol
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Alcohol
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05ts-5290000000-235e78c9d740ee21b7cf | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-00di-1110169000-0362236d3ab9846457e5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-64b022d4fccc5f107a63 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-053l-8920000000-1d2c2b37a5326eb8cca3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-1900000000-94593e549bc35d8f7d40 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a7l-0890000000-67d9a230a9e49dc395b4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9610000000-bac4b8dd8bd6ce35738b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-18a2a0e5278d3469af12 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0190000000-ab3d9485c704c7f50fc4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-1950000000-bf8ed52ffd013dc583fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4j-9820000000-97b9b0c9bc773e2e5b66 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-056r-1190000000-7cedee041402b0ddcc6a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-8940000000-e6d8221930258d6dbdc8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-78d3c140e7204d692961 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|