| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:27:12 UTC |
|---|
| Update Date | 2020-05-21 16:27:12 UTC |
|---|
| BMDB ID | BMDB0000238 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Sepiapterin |
|---|
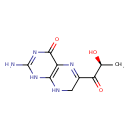
| Description | Sepiapterin, also known as lopac-S-154, belongs to the class of organic compounds known as pterins and derivatives. These are polycyclic aromatic compounds containing a pterin moiety, which consist of a pteridine ring bearing a ketone and an amine group to form 2-aminopteridin-4(3H)-one. Sepiapterin is an extremely weak basic (essentially neutral) compound (based on its pKa). Sepiapterin is a potentially toxic compound. Sepiapterin, with regard to humans, has been linked to the inborn metabolic disorder sepiapterin reductase deficiency. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2-Amino-7,8-dihydro-6-(2-hydroxy-1-oxopropyl)-4(1H)-pteridinone | HMDB | | 1-(2-Amino-7,8-dihydro-4-hydroxy-6-pteridinyl)-2-hydroxy-1-propanone | HMDB | | 2-Amino-6-(S)-lactoyl-7,8-dihydro-4(3H)-pteridinone | HMDB | | 2-Amino-7,8-dihydro-6-[(2S)-2-hydroxy-1-oxopropyl]-4(1H)pteridinone | HMDB | | L-Sepiapterin | HMDB | | Lopac-S-154 | HMDB | | Sepiapterine | HMDB | | Sepiapterin-C | HMDB | | 2-Amino-6-(S)-lactoyl-7,8-dihydro-4(3H)- pteridinone | HMDB | | Sepia-pterin | HMDB | | 2-Amino-7,8-dihydro-6-((2S)-2-hydroxy-1-oxopropyl)-4(3H)-pteridinone acid | HMDB |
|
|---|
| Chemical Formula | C9H11N5O3 |
|---|
| Average Molecular Weight | 237.2153 |
|---|
| Monoisotopic Molecular Weight | 237.086189243 |
|---|
| IUPAC Name | 2-amino-6-[(2S)-2-hydroxypropanoyl]-1,4,7,8-tetrahydropteridin-4-one |
|---|
| Traditional Name | sepiapterin |
|---|
| CAS Registry Number | 17094-01-8 |
|---|
| SMILES | C[C@H](O)C(=O)C1=NC2=C(NC1)NC(N)=NC2=O |
|---|
| InChI Identifier | InChI=1S/C9H11N5O3/c1-3(15)6(16)4-2-11-7-5(12-4)8(17)14-9(10)13-7/h3,15H,2H2,1H3,(H4,10,11,13,14,17)/t3-/m0/s1 |
|---|
| InChI Key | VPVOXUSPXFPWBN-VKHMYHEASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pterins and derivatives. These are polycyclic aromatic compounds containing a pterin moiety, which consist of a pteridine ring bearing a ketone and an amine group to form 2-aminopteridin-4(3H)-one. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Pterins and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pterin
- Aminopyrimidine
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Acyloin
- Pyrimidine
- Alpha-hydroxy ketone
- Heteroaromatic compound
- Vinylogous amide
- Secondary alcohol
- Ketimine
- Ketone
- Propargyl-type 1,3-dipolar organic compound
- Secondary amine
- Azacycle
- Organic 1,3-dipolar compound
- Alcohol
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxide
- Imine
- Carbonyl group
- Organic oxygen compound
- Amine
- Hydrocarbon derivative
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03di-5910000000-428f8e46f66f7bfd3119 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-000j-6590000000-f5711e35dabd2148a990 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00ko-0950000000-2cf5caf622d639c89359 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0297-2900000000-f9fe1627a36e71b61db6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00di-3900000000-82cabe6461c502831c78 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0079-0090000000-1e9c5af672b8662896af | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0940000000-1e5701cfd0eed57e783e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03k9-2910000000-3e376e485f9ccca285f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0290000000-9eff7a7965f178fa7e5e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-1920000000-a634b611cb12a5f44b6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-9200000000-ddd484b0e0f2ed7d6cd7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0090000000-f2b1b9dcb9b57fb43f71 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00ri-0490000000-28ee82ba7ca76d1a742b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6t-4900000000-741cb1d476965a4e887f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000l-0970000000-ade13f4ae6be4ca0759d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ox-0900000000-1fc5e869af92f5737ea4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9500000000-d928f0c83deb69c4394c | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|