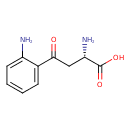
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-014l-0931000000-29fa7c323b035cb399d3 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014l-0931000000-29fa7c323b035cb399d3 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014l-0931000000-a43ae0acafac266101c4 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0006-1910000000-fdc25d09b4da4b1d7657 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00dl-9500000000-d7bf6a26c74e0dcabf40 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-4920000000-cb575b96b16b3ef9130f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0006-2910000000-366ce08b5736d68dfad0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-01bd-5900000000-c18e10be7e029474cf03 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00kf-9300000000-0f8238d38819f6f30d12 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0a4i-0790000000-3fa45266412288bf1c62 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0007-3900000000-a66f45aff81960af7575 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00xv-5900000000-15442ec5aaef005401ae | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-00r7-7900000000-0833fe9d2739d329f197 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0006-9400000000-d5d45364462a730de436 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-0006-0900000000-13929f0bde71b6326d3f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-0f6w-2900000000-b2d4f361ffee9fd93bb7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-004j-0900000000-96930a886f7bbcc44df3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-000i-0900000000-2441a358973abdb063da | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-0udi-0900000000-dc4fee4751d629575157 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0006-0900000000-4ec06cc06a3eab2625d6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0006-0900000000-319acefac99779fa7552 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0059-0890000000-4280b92b875a6f754a91 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0059-0890000000-664e2c8904363418682e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-00dj-0139000000-aa6188977ba2f460f5bc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-00dj-0139000000-ee71a364df13bd095feb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06r7-0920000000-f2e6efdd3988d3010e33 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-022a-3900000000-e1c06099462847ad5b97 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9700000000-a9df2d5ca325f97f69f1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-2490000000-9604ae0cc9d437acd878 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dl-9520000000-8f0ea7d174290496860d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-9400000000-9721076505654380d2de | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|