| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:39:29 UTC |
|---|
| Update Date | 2020-05-21 16:28:44 UTC |
|---|
| BMDB ID | BMDB0000985 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Dihydrolipoamide |
|---|
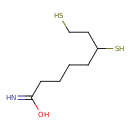
| Description | Dihydrolipoamide belongs to the class of organic compounds known as fatty amides. These are carboxylic acid amide derivatives of fatty acids, that are formed from a fatty acid and an amine. Thus, dihydrolipoamide is considered to be a fatty amide lipid molecule. Dihydrolipoamide is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Dihydrolipoamide exists in all living species, ranging from bacteria to humans. Dihydrolipoamide participates in a number of enzymatic reactions, within cattle. In particular, Dihydrolipoamide can be converted into lipoamide; which is catalyzed by the enzyme dihydrolipoyl dehydrogenase E3. In addition, Dihydrolipoamide can be biosynthesized from S-(3-methylbutanoyl)-dihydrolipoamide-e; which is mediated by the enzyme lipoamide acyltransferase component OF branched-chain alpha-keto acid dehydrogenase complex, mitochondrial. In cattle, dihydrolipoamide is involved in the metabolic pathway called the valine, leucine, and isoleucine degradation pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6,8-Dimercaptooctanamide | ChEBI | | Dihydrothioctamide | ChEBI | | 6,8-Bis-sulfanyloctanamide | HMDB | | 6,8-Dimercapto-octanamide | HMDB | | 6,8-Disulfanyloctanamide | HMDB | | Dihydrolipoamide, (+-)-isomer | HMDB |
|
|---|
| Chemical Formula | C8H17NOS2 |
|---|
| Average Molecular Weight | 207.357 |
|---|
| Monoisotopic Molecular Weight | 207.075155551 |
|---|
| IUPAC Name | 6,8-disulfanyloctanimidic acid |
|---|
| Traditional Name | dihydrothioctamide |
|---|
| CAS Registry Number | 3884-47-7 |
|---|
| SMILES | OC(=N)CCCCC(S)CCS |
|---|
| InChI Identifier | InChI=1S/C8H17NOS2/c9-8(10)4-2-1-3-7(12)5-6-11/h7,11-12H,1-6H2,(H2,9,10) |
|---|
| InChI Key | VLYUGYAKYZETRF-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty amides. These are carboxylic acid amide derivatives of fatty acids, that are formed from a fatty acid and an amine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty amides |
|---|
| Direct Parent | Fatty amides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty amide
- Primary carboxylic acid amide
- Carboxamide group
- Carboxylic acid derivative
- Alkylthiol
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Mitochondria
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9700000000-936a7170a2da09aecf11 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9410000000-9f1410bc40e59db39d21 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4l-0940000000-f34746a5ea022152ba75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052f-1900000000-99beaa28d55837788743 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0m2i-9400000000-53152f2f29c03cd0cbcd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ab9-1960000000-1e45a79f64407fb4dea8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ab9-5930000000-297b157da8ed63567faa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-82e7dacb48c8cab103d4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ab9-0950000000-6272c55454b18f718c5d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0w29-2900000000-a19052123ea01afd8613 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002v-9300000000-c0ae302dda03ee321d60 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-1361b5d8ba6f1d1a13ef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0pb9-5940000000-00e811c07c89a8896c7d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-90726b17dc36e29c5299 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|