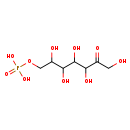| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:40:42 UTC |
|---|
| Update Date | 2020-05-11 20:38:37 UTC |
|---|
| BMDB ID | BMDB0001068 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | D-Sedoheptulose 7-phosphate |
|---|
| Description | D-D-d-sedoheptulose 7-phosphate, also known as D-sedoheptulose-7-P, belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. D-D-d-sedoheptulose 7-phosphate is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). D-D-d-sedoheptulose 7-phosphate exists in all eukaryotes, ranging from yeast to humans. D-D-d-sedoheptulose 7-phosphate participates in a number of enzymatic reactions, within cattle. In particular, D-Glyceraldehyde 3-phosphate and D-d-sedoheptulose 7-phosphate can be converted into D-ribose 5-phosphate and xylulose 5-phosphate through its interaction with the enzyme transketolase. Furthermore, D-Glyceraldehyde 3-phosphate and D-d-sedoheptulose 7-phosphate can be biosynthesized from D-erythrose 4-phosphate and fructose 6-phosphate; which is mediated by the enzyme transaldolase. Furthermore, D-Glyceraldehyde 3-phosphate and D-d-sedoheptulose 7-phosphate can be biosynthesized from D-erythrose 4-phosphate and fructose 6-phosphate through its interaction with the enzyme transaldolase. Finally, D-Glyceraldehyde 3-phosphate and D-d-sedoheptulose 7-phosphate can be converted into D-ribose 5-phosphate and xylulose 5-phosphate; which is catalyzed by the enzyme transketolase. In cattle, D-d-sedoheptulose 7-phosphate is involved in a couple of metabolic pathways, which include the pentose phosphate pathway and cancer (via the Warburg effect). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| D-Sedoheptulose 7-phosphoric acid | Generator | | Sedoheptulose 7-phosphate | MeSH, HMDB | | Heptulose-7-phosphate | MeSH, HMDB | | Sedoheptulose 7-phosphoric acid(2-) | Generator |
|
|---|
| Chemical Formula | C7H15O10P |
|---|
| Average Molecular Weight | 290.1618 |
|---|
| Monoisotopic Molecular Weight | 290.040283212 |
|---|
| IUPAC Name | [(2,3,4,5,7-pentahydroxy-6-oxoheptyl)oxy]phosphonic acid |
|---|
| Traditional Name | sedoheptulose 7-phosphate |
|---|
| CAS Registry Number | 2646-35-7 |
|---|
| SMILES | OCC(=O)C(O)C(O)C(O)C(O)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C7H15O10P/c8-1-3(9)5(11)7(13)6(12)4(10)2-17-18(14,15)16/h4-8,10-13H,1-2H2,(H2,14,15,16) |
|---|
| InChI Key | JDTUMPKOJBQPKX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Monosaccharide phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Heptose monosaccharide
- Monosaccharide phosphate
- Monoalkyl phosphate
- Acyloin
- Beta-hydroxy ketone
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Alpha-hydroxy ketone
- Ketone
- Secondary alcohol
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Primary alcohol
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0532-9010163000-acf4f0352711d42c5fd6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-3590000000-b34117115db1142c20a4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4m-9710000000-f24eb76da4873de6d41f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9300000000-b8503ab37c97a6fff189 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-052r-9730000000-02687371ce694d179c1a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9200000000-27d52a9bf7e9b234f147 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4a6e471e293da809c157 | View in MoNA |
|---|
|
|---|