| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:41:40 UTC |
|---|
| Update Date | 2020-05-21 16:29:06 UTC |
|---|
| BMDB ID | BMDB0001143 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
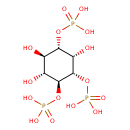
| Common Name | Inositol 1,3,4-trisphosphate |
|---|
| Description | Inositol 1,3,4-trisphosphate, also known as 1,3,4-itp or I3S, belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. Inositol 1,3,4-trisphosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Inositol 1,3,4-trisphosphate exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (1S,3S,4S)-1,3,4-TRIPHOSPHO-myo-inositol | ChEBI | | D-Myo-inositol 1,3,4-trisphosphate | ChEBI | | D-Myo-inositol 1,3,4-trisphosphoric acid | Generator | | Inositol 1,3,4-trisphosphoric acid | Generator | | 1,3,4-Itp | HMDB | | 1D-Myo-inositol 1,3,4-trisphosphate | HMDB | | I3S | HMDB | | Inositol 1,3,4-triphosphate | HMDB | | Myo-inositol 1,3,4-trisphosphate | HMDB | | Inositol 1,3,4-trisphosphate, (D)-isomer | HMDB | | D-myo-Inositol 1,3,4-tris(phosphate) | HMDB | | Inositol 1,3,4-trisphosphate | HMDB | | Ins(1,3,4)P3 | HMDB |
|
|---|
| Chemical Formula | C6H15O15P3 |
|---|
| Average Molecular Weight | 420.0956 |
|---|
| Monoisotopic Molecular Weight | 419.962379346 |
|---|
| IUPAC Name | {[(1S,2R,3R,4S,5S,6S)-2,3,5-trihydroxy-4,6-bis(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | inositol 1,3,4-trisphosphate |
|---|
| CAS Registry Number | 98102-63-7 |
|---|
| SMILES | O[C@@H]1[C@@H](O)[C@H](OP(O)(O)=O)[C@@H](OP(O)(O)=O)[C@@H](O)[C@H]1OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H15O15P3/c7-1-2(8)5(20-23(13,14)15)6(21-24(16,17)18)3(9)4(1)19-22(10,11)12/h1-9H,(H2,10,11,12)(H2,13,14,15)(H2,16,17,18)/t1-,2-,3+,4+,5+,6+/m1/s1 |
|---|
| InChI Key | MMWCIQZXVOZEGG-MLQGYMEPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Monoalkyl phosphate
- Cyclohexanol
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002b-7195200000-2871156f7c1e00704088 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0fka-8912544000-6466de288b248e0534c9 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0006-0091000000-4748b18c24a28a83e10b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-00ba-9030000000-32edb6df84b89249800a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, negative | splash10-0udi-0000900000-1b3fca9ae09a6e9974b5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 13V, negative | splash10-0udi-0000900000-a276fc4491e761718dfa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 16V, negative | splash10-0udi-0002900000-bcbe8de3f6c9f3878dc2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 21V, negative | splash10-0udi-1609700000-8524975955b3b52a5188 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 26V, negative | splash10-0pk9-4809100000-bd4057c77d97fcd8899c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 31V, negative | splash10-056r-9805000000-7df4af8b6064aead614d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 36V, negative | splash10-004i-9401000000-4c1e912ddb150b70b5d4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 43V, negative | splash10-004i-9200000000-84c539d19c53654b7250 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 50V, negative | splash10-004i-9100000000-588efa0963cb71999431 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 60V, negative | splash10-004i-9000000000-6a2554d172da41062ddd | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0udi-0109000000-0e68666378959fa09055 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-004i-9000000000-46d4cee1b5ac630ba9b8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0zfr-0890000000-107341de028331b689c2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-004i-6900000000-dc0e986525b75ecb2fb0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-08gi-0849000000-19971dee01d98c786cd0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0a4i-0952000000-fd8d641519350c7213ea | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, negative | splash10-014i-0000900000-d3ffebcbd2b7887c7769 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-3008900000-e987cfb2603d94f1a6a4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uk9-2006900000-95047338604f8928313f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-4291000000-8e9bb388f01ec6d535ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-4001900000-395b5e207aa26dd43f27 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9003200000-5e6369906fdedd08ad60 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4b72b937fe3bf7fe817e | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|