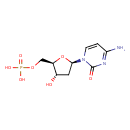| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:33 UTC |
|---|
| Update Date | 2020-05-21 16:29:01 UTC |
|---|
| BMDB ID | BMDB0001202 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | dCMP |
|---|
| Description | dCMP, also known as deoxycytidylate, belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside monophosphates. These are pyrimidine nucleotides with a monophosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. dCMP is an extremely weak basic (essentially neutral) compound (based on its pKa). dCMP exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2'-Deoxycytidine 5'-monophosphate | ChEBI | | 2'-DEOXYCYTIDINE-5'-monophosphATE | ChEBI | | Deoxycytidine monophosphate | ChEBI | | Deoxycytidylate | ChEBI | | Deoxycytidylic acid | ChEBI | | 2'-Deoxycytidine 5'-monophosphoric acid | Generator | | 2'-DEOXYCYTIDINE-5'-monophosphoric acid | Generator | | Deoxycytidine monophosphoric acid | Generator | | 2'-Deoxycytosine 5'-monophosphate | HMDB | | 2'-Deoxycytosine 5'-monophosphoric acid | HMDB | | 2'-Deoxycytidine-3'-monophosphate | HMDB | | 2'-Deoxycytidine-5'-monophosphorate | HMDB | | 2'-Deoxycytidine-5'-phosphate | HMDB | | Deoxycytidine-phosphate | HMDB | | Acid, deoxycytidylic | HMDB | | Deoxycytidylic acids | HMDB | | Acids, deoxycytidylic | HMDB | | monoPhosphate, deoxycytidine | HMDB |
|
|---|
| Chemical Formula | C9H14N3O7P |
|---|
| Average Molecular Weight | 307.1971 |
|---|
| Monoisotopic Molecular Weight | 307.056936329 |
|---|
| IUPAC Name | {[(2R,3S,5R)-5-(4-amino-2-oxo-1,2-dihydropyrimidin-1-yl)-3-hydroxyoxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | dCMP |
|---|
| CAS Registry Number | 1032-65-1 |
|---|
| SMILES | NC1=NC(=O)N(C=C1)[C@H]1C[C@H](O)[C@@H](COP(O)(O)=O)O1 |
|---|
| InChI Identifier | InChI=1S/C9H14N3O7P/c10-7-1-2-12(9(14)11-7)8-3-5(13)6(19-8)4-18-20(15,16)17/h1-2,5-6,8,13H,3-4H2,(H2,10,11,14)(H2,15,16,17)/t5-,6+,8+/m0/s1 |
|---|
| InChI Key | NCMVOABPESMRCP-SHYZEUOFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside monophosphates. These are pyrimidine nucleotides with a monophosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine deoxyribonucleotides |
|---|
| Direct Parent | Pyrimidine 2'-deoxyribonucleoside monophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine 2'-deoxyribonucleoside monophosphate
- Aminopyrimidine
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Imidolactam
- Alkyl phosphate
- Heteroaromatic compound
- Tetrahydrofuran
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Primary amine
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Amine
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cytoplasm
- Lysosome
- Mitochondria
- Nucleus
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9410000000-f31bf162f5f972b9cf42 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-03dm-9522000000-d25b21eb547d23c4886d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-0900000000-01bb64efeca205daaf6e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03di-1900000000-13c56ec900e188e10ddb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-03di-5900000000-4e6dd8fdc8a4142269a2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-004j-9402000000-6a9e4b2e32058773f6bf | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 35V, negative | splash10-0006-9000000000-3b4572f2c8e51186f7e5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-bc40821757dbbe57a7c2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-1900000000-9052cfbf31794c457d3b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004j-9400000000-1f2a185e05aed001e983 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-5209000000-b46ae8ade9397163198a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9300000000-c6505ac4fafc2bc54910 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-2d1a73d09ad4b06da15f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-03fr-3910000000-ee0cc0c7897818415edc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-2900000000-7e33f3b263e35a18c2e0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03dj-9400000000-4ab96ba8107b5f1820a6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-3900000000-56a3ecce668ff56b702c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0900000000-986f256cdcdf8c8d2f9f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0006-9000000000-d236c094fbaf8d2136b4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-dd904e0c162b36055113 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-4cc5b5143e38514f3010 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-3b0a0a6948a49378ff1c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-4900000000-a901f489d4e9b8516bf2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-6900000000-d9e59981c244792a2f76 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6r-9367000000-d5c1062e5203ec9699d4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9110000000-20768d2b834097ac11d7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-29f2d493f3f68928488b | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|