| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:45 UTC |
|---|
| Update Date | 2020-05-21 16:29:01 UTC |
|---|
| BMDB ID | BMDB0001227 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5-Thymidylic acid |
|---|
| Description | 5-Thymidylic acid, also known as TMP or thymidylate, belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside monophosphates. These are pyrimidine nucleotides with a monophosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. 5-Thymidylic acid is an extremely weak basic (essentially neutral) compound (based on its pKa). 5-Thymidylic acid exists in all living species, ranging from bacteria to humans. Within cattle, 5-thymidylic acid participates in a number of enzymatic reactions. In particular, 5-thymidylic acid and dihydrofolic acid can be biosynthesized from dUMP and 5,10-methylene-THF; which is mediated by the enzyme thymidylate synthase. In addition, 5-thymidylic acid can be converted into dTDP through its interaction with the enzyme thymidylate synthase. In cattle, 5-thymidylic acid is involved in the metabolic pathway called the pyrimidine metabolism pathway. |
|---|
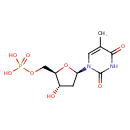
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (DT)1 | ChEBI | | 5'-Thymidylic acid | ChEBI | | 5'-TMP | ChEBI | | 5-Methyl-dUMP | ChEBI | | Deoxyribosylthymine monophosphate | ChEBI | | Ribothymidine 5'-monophosphate | ChEBI | | Thymidine 5'-(dihydrogen phosphate) | ChEBI | | Thymidine 5'-phosphate | ChEBI | | Thymidine 5'-phosphoric acid | ChEBI | | Thymidine monophosphate | ChEBI | | Thymidine-5'-monophosphoric acid | ChEBI | | THYMIDINE-5'-phosphATE | ChEBI | | Thymidylate | ChEBI | | Thymidylic acid | ChEBI | | TMP | ChEBI | | Deoxythymidine 5'-phosphate | Kegg | | Deoxythymidylic acid | Kegg | | 5'-Thymidylate | Generator | | Deoxyribosylthymine monophosphoric acid | Generator | | Ribothymidine 5'-monophosphoric acid | Generator | | Thymidine 5'-(dihydrogen phosphoric acid) | Generator | | Thymidine monophosphoric acid | Generator | | Thymidine-5'-monophosphate | Generator | | THYMIDINE-5'-phosphoric acid | Generator | | Deoxythymidine 5'-phosphoric acid | Generator | | Deoxythymidylate | Generator | | 5-Thymidylate | Generator | | 2'-Deoxythymidine 5'-monophosphate | HMDB | | 5'-dTMP | HMDB | | Deoxy TMP | HMDB | | Deoxythymidine 5'-monophosphate | HMDB | | Deoxythymidine monophosphate | HMDB | | Deoxythymydilate | HMDB | | Deoxythymydilic acid | HMDB | | dTMP | HMDB | | Thymidine 5'-monophosphate | HMDB | | Thymidine 5'-phosphorate | HMDB | | Thymidine 5'MP | HMDB | | Thymidine mononucleotide | HMDB | | Thymidine phosphate | HMDB | | Thymidine-5'-monophosphorate | HMDB | | Thymidylic acids | HMDB | | Acids, thymidylic | HMDB | | Acid, thymidylic | HMDB | | monoPhosphate, thymidine | HMDB |
|
|---|
| Chemical Formula | C10H15N2O8P |
|---|
| Average Molecular Weight | 322.2085 |
|---|
| Monoisotopic Molecular Weight | 322.056601978 |
|---|
| IUPAC Name | {[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)oxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | thymidylate |
|---|
| CAS Registry Number | 365-07-1 |
|---|
| SMILES | CC1=CN([C@H]2C[C@H](O)[C@@H](COP(O)(O)=O)O2)C(=O)NC1=O |
|---|
| InChI Identifier | InChI=1S/C10H15N2O8P/c1-5-3-12(10(15)11-9(5)14)8-2-6(13)7(20-8)4-19-21(16,17)18/h3,6-8,13H,2,4H2,1H3,(H,11,14,15)(H2,16,17,18)/t6-,7+,8+/m0/s1 |
|---|
| InChI Key | GYOZYWVXFNDGLU-XLPZGREQSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside monophosphates. These are pyrimidine nucleotides with a monophosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine deoxyribonucleotides |
|---|
| Direct Parent | Pyrimidine 2'-deoxyribonucleoside monophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine 2'-deoxyribonucleoside monophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Lactam
- Secondary alcohol
- Urea
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cytoplasm
- Lysosome
- Mitochondria
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-01u0-0961000000-4380a23f8bec3c2dd015 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-03k9-0940000000-6238904f1b1f293aaf3a | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-01u0-0961000000-4380a23f8bec3c2dd015 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-03k9-0940000000-6238904f1b1f293aaf3a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9500000000-ca4d82d816839360b115 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0002-9621000000-1a0c588cd8f192b77565 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-9110000000-a45c0d5a58cdb0e5fee4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-624fe22ca203d0e6430e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9000000000-0c23943cc868acaaeda4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-004i-9703000000-452b674ca61adb40209b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-004i-9703000000-452b674ca61adb40209b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9000000000-2233c579a23110abe895 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9000000000-9af792986db292f0d8d1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9010000000-9936ff7b595382418b7c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9200000000-5c7a9e2c6c970eec2d92 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9000000000-edfd7498bf3ebaedbb4b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9000000000-9dafb0586b1aadb8475d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-001i-9100000000-b286ae95b38725486f5b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9000000000-c716e9bc2009f1adb3ae | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-ec3ffc3673cc769d3a0e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004i-8900000000-90b7e520ce562f39a280 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004i-7900000000-a01024a3e21d2de7f133 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004j-5900000000-57620f7a2b0abf78c220 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-004i-8900000000-b0b0ac82c08ccb2ea84e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-006t-0905000000-49f5a9d8a112c369032d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-b932c025cbc139ba930b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-4900000000-ae144ce0df705b85f237 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-5900000000-9f2e5ba96fb24ff28b7f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9766000000-855dcb81a8d160218e77 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9200000000-f9116de370dd3211ece6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-0953b010bfa0e123afa7 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|