| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:44:55 UTC |
|---|
| Update Date | 2020-05-21 16:28:34 UTC |
|---|
| BMDB ID | BMDB0001373 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Dephospho-CoA |
|---|
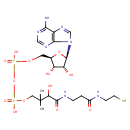
| Description | Dephospho-CoA, also known as Dephospho-coa, belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. Thus, dephospho-CoA is considered to be a fatty ester lipid molecule. Dephospho-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Dephospho-CoA exists in all living species, ranging from bacteria to humans. In cattle, dephospho-CoA is involved in the metabolic pathway called pantothenate and CoA biosynthesis pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3'-Dephospho-CoA | HMDB | | 3'-Dephospho-coenzyme A | HMDB | | 3'-O-Dephosphono-CoA | HMDB | | 3'-O-Dephosphono-coenzyme A | HMDB | | Dephospho-coenzyme A | HMDB | | Dephosphocoenzyme A | HMDB | | DpCoA | MeSH | | Dephospho CoA | MeSH | | 3'-Desphospho-coenzyme A | MeSH | | 2'-(5''-Triphosphoribosyl)-3'-dephospho-CoA | MeSH | | Desphospho-CoA | MeSH |
|
|---|
| Chemical Formula | C21H35N7O13P2S |
|---|
| Average Molecular Weight | 687.554 |
|---|
| Monoisotopic Molecular Weight | 687.148877955 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][3-hydroxy-2,2-dimethyl-3-({2-[(2-sulfanylethyl)carbamoyl]ethyl}carbamoyl)propoxy]phosphinic acid |
|---|
| Traditional Name | {[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(3-hydroxy-2,2-dimethyl-3-({2-[(2-sulfanylethyl)carbamoyl]ethyl}carbamoyl)propoxy)phosphinic acid |
|---|
| CAS Registry Number | 3633-59-8 |
|---|
| SMILES | CC(C)(COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC2=C1N=CN=C2N)C(O)C(=O)NCCC(=O)NCCS |
|---|
| InChI Identifier | InChI=1S/C21H35N7O13P2S/c1-21(2,16(32)19(33)24-4-3-12(29)23-5-6-44)8-39-43(36,37)41-42(34,35)38-7-11-14(30)15(31)20(40-11)28-10-27-13-17(22)25-9-26-18(13)28/h9-11,14-16,20,30-32,44H,3-8H2,1-2H3,(H,23,29)(H,24,33)(H,34,35)(H,36,37)(H2,22,25,26)/t11-,14-,15-,16?,20-/m1/s1 |
|---|
| InChI Key | KDTSHFARGAKYJN-DRCCLKDXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Pentose monosaccharide
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Pyrimidine
- Fatty amide
- Imidolactam
- Monosaccharide
- Fatty acyl
- Phosphoric acid ester
- Alkyl phosphate
- Azole
- Heteroaromatic compound
- Imidazole
- Tetrahydrofuran
- Secondary alcohol
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Organoheterocyclic compound
- Carboxylic acid derivative
- Alkylthiol
- Oxacycle
- Azacycle
- Organic oxide
- Organooxygen compound
- Organosulfur compound
- Carbonyl group
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Alcohol
- Organonitrogen compound
- Primary amine
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03fr-1452933000-479a35a051c9c3fcda0d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-2930102000-19d2796dee5d14788e39 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-3930000000-d4541feacee9a6c7a4e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-4900000000-1307cbce80b215d981b9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0040-2910618000-58e100b1447a771ecd7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1910200000-ba57f132dee9798389fa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a7i-3900000000-d382c7ac6874be7ea151 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|