| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:46:31 UTC |
|---|
| Update Date | 2020-05-21 16:28:58 UTC |
|---|
| BMDB ID | BMDB0001480 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 3-Dehydrosphinganine |
|---|
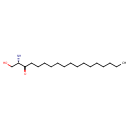
| Description | 3-Dehydrosphinganine, also known as KDHS or ketodihydrosphingosine, belongs to the class of organic compounds known as beta-hydroxy ketones. These are ketones containing a hydroxyl group attached to the beta-carbon atom, relative to the C=O group. Thus, 3-dehydrosphinganine is considered to be a sphingoid base lipid molecule. 3-Dehydrosphinganine is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. 3-Dehydrosphinganine exists in all eukaryotes, ranging from yeast to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Dehydro-D-sphinganine | ChEBI | | 3-Ketodihydrosphingosine | ChEBI | | 3-Ketosphinganine | ChEBI | | (2S)-2-Amino-1-hydroxyoctadecan-3-one | HMDB | | 1-Hydroxy-2-amino-3-oxo-octadecane | HMDB | | 2-Amino-1-hydroxy-3-octadecanone | HMDB | | KDHS | HMDB | | Ketodihydrosphingosine | HMDB | | (+-)-Isomer OF ketodihydrosphingosine | HMDB | | (S)-Isomer OF ketodihydrosphingosine | HMDB |
|
|---|
| Chemical Formula | C18H37NO2 |
|---|
| Average Molecular Weight | 299.4919 |
|---|
| Monoisotopic Molecular Weight | 299.282429433 |
|---|
| IUPAC Name | (2S)-2-amino-1-hydroxyoctadecan-3-one |
|---|
| Traditional Name | 3-ketosphinganine |
|---|
| CAS Registry Number | 16105-69-4 |
|---|
| SMILES | CCCCCCCCCCCCCCCC(=O)[C@@H](N)CO |
|---|
| InChI Identifier | InChI=1S/C18H37NO2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-18(21)17(19)16-20/h17,20H,2-16,19H2,1H3/t17-/m0/s1 |
|---|
| InChI Key | KBUNOSOGGAARKZ-KRWDZBQOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as beta-hydroxy ketones. These are ketones containing a hydroxyl group attached to the beta-carbon atom, relative to the C=O group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Beta-hydroxy ketones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta-hydroxy ketone
- Alpha-aminoketone
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Primary alcohol
- Organonitrogen compound
- Primary aliphatic amine
- Amine
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
- Mitochondria
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01p9-9120000000-135052b999433b5dd30b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-03di-9321000000-e0d96cb3bea3c320412a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-2096000000-8323a157b44b31f2331a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01qa-4491000000-39be7a6025f78d062cb5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ox-9510000000-a675e52903f069501f67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1090000000-e36f44a44812cc09023c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-015j-4090000000-4a24200105a7fd0afd1e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9020000000-edd2ec1e7a7fae113d2f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ue9-3069000000-e43a689cbba7742dc5c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-9282000000-f83e19a33602a8a25cb8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-f16a410d8c3330af51f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014j-0090000000-7d1c46d2460baf58b0dc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06ej-3090000000-b775508a0489f38a77f7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9010000000-b10c9eb2adca17092c8c | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|