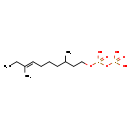
Showing metabocard for Dolichyl diphosphate (BMDB0001513)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-09-30 22:47:02 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2020-04-22 15:08:31 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BMDB ID | BMDB0001513 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Dolichyl diphosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Dolichyl diphosphate belongs to the class of organic compounds known as organic pyrophosphates. These are organic compounds containing the pyrophosphate oxoanion, with the structure OP([O-])(=O)OP(O)([O-])=O. Dolichyl diphosphate is a moderately acidic compound (based on its pKa). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C12H26O7P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 344.2782 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 344.11537621 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | [({[(7E)-3,8-dimethyldec-7-en-1-yl]oxy}(hydroxy)phosphoryl)oxy]phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | {[(7E)-3,8-dimethyldec-7-en-1-yl]oxy(hydroxy)phosphoryl}oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 37247-98-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CC\C(C)=C\CCCC(C)CCOP(O)(=O)OP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C12H26O7P2/c1-4-11(2)7-5-6-8-12(3)9-10-18-21(16,17)19-20(13,14)15/h7,12H,4-6,8-10H2,1-3H3,(H,16,17)(H2,13,14,15)/b11-7+ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | FFBTVOMODCZDNV-YRNVUSSQSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as organic pyrophosphates. These are organic compounds containing the pyrophosphate oxoanion, with the structure OP([O-])(=O)OP(O)([O-])=O. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organic oxoanionic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Organic pyrophosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Organic pyrophosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Expected but not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofunction | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Application | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0001513 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB022667 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 13748633 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | C00621 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | 6290 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | 5462166 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 15750 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in dolichyl-diphosphooligosaccharide-protein g
- Specific function:
- Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity.
- Gene Name:
- DAD1
- Uniprot ID:
- Q5E9C2
- Molecular weight:
- 12495.0
- General function:
- Involved in dolichyl-diphosphooligosaccharide-protein g
- Specific function:
- Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity (By similarity). Required for the assembly of both SST3A- and SS3B-containing OST complexes (By similarity).
- Gene Name:
- DDOST
- Uniprot ID:
- A6QPY0
- Molecular weight:
- 48791.0
- General function:
- Involved in dolichyl-diphosphooligosaccharide-protein g
- Specific function:
- Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity.
- Gene Name:
- RPN2
- Uniprot ID:
- Q3SZI6
- Molecular weight:
- 69214.0
- General function:
- Involved in dolichyl-diphosphooligosaccharide-protein g
- Specific function:
- Catalytic subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity. This subunit contains the active site and the acceptor peptide and donor lipid-linked oligosaccharide (LLO) binding pockets (By similarity). STT3A is present in the majority of OST complexes and mediates cotranslational N-glycosylation of most sites on target proteins, while STT3B-containing complexes are required for efficient post-translational glycosylation and mediate glycosylation of sites that have been skipped by STT3A (By similarity).
- Gene Name:
- STT3A
- Uniprot ID:
- Q2KJI2
- Molecular weight:
- 80656.0