| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:50:40 UTC |
|---|
| Update Date | 2020-05-21 16:29:04 UTC |
|---|
| BMDB ID | BMDB0002065 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 6-Lactoyltetrahydropterin |
|---|
| Description | 6-Lactoyltetrahydropterin belongs to the class of organic compounds known as pterins and derivatives. These are polycyclic aromatic compounds containing a pterin moiety, which consist of a pteridine ring bearing a ketone and an amine group to form 2-aminopteridin-4(3H)-one. 6-Lactoyltetrahydropterin exists in all living organisms, ranging from bacteria to humans. Based on a literature review a small amount of articles have been published on 6-Lactoyltetrahydropterin. |
|---|
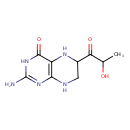
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Amino-6-lactoyl-5,6,7,8-tetrahydropteridin-4(3H)-one | ChEBI | | 6-1'-oxo-2'-Hydroxypropyl tetrahydropterin | ChEBI | | 6-Lactoyl tetrahydropterin | HMDB | | 6-Lactoyl-5,6,7,8-tetrahydropterin | HMDB | | Lactoyl-H4-pterin | HMDB | | Lactoyl-PH4 | HMDB | | 1'-oxo-2'-Hydroxypropyl-tetrahydropterin | MeSH, HMDB |
|
|---|
| Chemical Formula | C9H13N5O3 |
|---|
| Average Molecular Weight | 239.2312 |
|---|
| Monoisotopic Molecular Weight | 239.101839307 |
|---|
| IUPAC Name | 2-amino-6-(2-hydroxypropanoyl)-3,4,5,6,7,8-hexahydropteridin-4-one |
|---|
| Traditional Name | 6-lactoyl tetrahydropterin |
|---|
| CAS Registry Number | 33405-80-0 |
|---|
| SMILES | CC(O)C(=O)C1CNC2=C(N1)C(=O)NC(N)=N2 |
|---|
| InChI Identifier | InChI=1S/C9H13N5O3/c1-3(15)6(16)4-2-11-7-5(12-4)8(17)14-9(10)13-7/h3-4,12,15H,2H2,1H3,(H4,10,11,13,14,17) |
|---|
| InChI Key | HKCYZTKHPLJZDR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pterins and derivatives. These are polycyclic aromatic compounds containing a pterin moiety, which consist of a pteridine ring bearing a ketone and an amine group to form 2-aminopteridin-4(3H)-one. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Pterins and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pterin
- Aminopyrimidine
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Acyloin
- Pyrimidine
- Alpha-aminoketone
- Heteroaromatic compound
- Vinylogous amide
- Alpha-hydroxy ketone
- 1,3-aminoalcohol
- Secondary alcohol
- Ketone
- Azacycle
- Secondary amine
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Primary amine
- Carbonyl group
- Organic nitrogen compound
- Amine
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-014i-2900000000-49dec0d6931d1e9bad0a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-014i-2900000000-8f1d69e548d027d65a19 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-0090000000-a0f9d3c66da5b6d5f581 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0940000000-176c06edb35b9da14cbc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0kmi-4910000000-a4fc7ca72b4b46138d0f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0290000000-6c319e8337df67fc476d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-1920000000-bfd50799ec0cb51b89fe | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fdo-9200000000-fe4583b38b03564abe97 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0190000000-57a11b44e9df0fc1d6a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00xr-0690000000-b7a86bbaa7a707429283 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002f-7900000000-0cb0aca82f0fb03cb603 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00kf-0910000000-77790fad0fe8dd8e8a4e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kf-0900000000-0d38a13d09e7cb8e4809 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9500000000-81ba6afc34a8818ef29d | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|