| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:53:57 UTC |
|---|
| Update Date | 2020-05-21 16:28:45 UTC |
|---|
| BMDB ID | BMDB0002310 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | S-Methylmalonyl-CoA |
|---|
| Description | S-Methylmalonyl-CoA belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. Based on a literature review very few articles have been published on S-Methylmalonyl-CoA. |
|---|
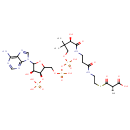
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-Methylmalonyl-CoA | HMDB | | (S)-Methylmalonyl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C25H40N7O19P3S |
|---|
| Average Molecular Weight | 867.607 |
|---|
| Monoisotopic Molecular Weight | 867.131252359 |
|---|
| IUPAC Name | (2S)-3-[(2-{3-[(2R)-3-[({[({[(3S,4R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-2-methyl-3-oxopropanoic acid |
|---|
| Traditional Name | (2S)-3-[(2-{3-[(2R)-3-{[({[(3S,4R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]methyl}-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-2-methyl-3-oxopropanoic acid |
|---|
| CAS Registry Number | 73173-91-8 |
|---|
| SMILES | C[C@@H](C(O)=O)C(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OCC1OC([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 |
|---|
| InChI Identifier | InChI=1S/C25H40N7O19P3S/c1-12(23(37)38)24(39)55-7-6-27-14(33)4-5-28-21(36)18(35)25(2,3)9-48-54(45,46)51-53(43,44)47-8-13-17(50-52(40,41)42)16(34)22(49-13)32-11-31-15-19(26)29-10-30-20(15)32/h10-13,16-18,22,34-35H,4-9H2,1-3H3,(H,27,33)(H,28,36)(H,37,38)(H,43,44)(H,45,46)(H2,26,29,30)(H2,40,41,42)/t12-,13?,16+,17+,18-,22?/m0/s1 |
|---|
| InChI Key | MZFOKIKEPGUZEN-JDVCRUKVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Alkyl phosphate
- 1,3-dicarbonyl compound
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Heteroaromatic compound
- Azole
- Thiocarboxylic acid ester
- Carbothioic s-ester
- Secondary alcohol
- Amino acid
- Carboxamide group
- Amino acid or derivatives
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organosulfur compound
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
- Mitochondria
- Peroxisome
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-4911000020-c392edc35bfe7a605c16 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-1913000000-d7d73fa71a22c4b9e10c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2911000000-1c3ee05de5b5cf1c067c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00nb-9811140550-fade85d2dd723fe43fda | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-5911110010-2c7ce12039a0e299c300 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a7i-5900100000-48f1904d8b30fe71d5e4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0000000190-f076790ab423a5aa6698 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gc0-1301103960-0fda0df24a3a3ad7fe59 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-1159000000-2510a377524f6c35c5f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000000490-c603c70c007f70ef33b6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9100001110-8a105fab331fa01db259 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f9b-6003404900-356070eb7375b08b8e14 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Mutka, Sarah C.; Bondi, Shana M.; Carney, John R.; Da Silva, Nancy A.; Kealey, James T. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae .FEMS Yeast Research (2006), 6(1), 40-47. |
|---|