| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:07:09 UTC |
|---|
| Update Date | 2020-05-21 16:29:05 UTC |
|---|
| BMDB ID | BMDB0003971 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Beta-D-Fructose 6-phosphate |
|---|
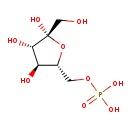
| Description | Beta-D-Fructose 6-phosphate, also known as b-D-fructose 6-phosphoric acid or 6-O-phosphono-b-D-fructofuranose, belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. Beta-D-Fructose 6-phosphate is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). Beta-D-Fructose 6-phosphate exists in all living species, ranging from bacteria to humans. Beta-D-Fructose 6-phosphate participates in a number of enzymatic reactions, within cattle. In particular, Beta-D-Fructose 6-phosphate can be biosynthesized from D-fructose through the action of the enzyme hexokinase-2. In addition, Beta-D-Fructose 6-phosphate can be converted into glucose 6-phosphate through its interaction with the enzyme glucose-6-phosphate isomerase. In cattle, Beta-D-fructose 6-phosphate is involved in the metabolic pathway called the starch and sucrose metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6-O-Phosphono-beta-D-fructofuranose | ChEBI | | FRUCTOSE-6-phosphATE | ChEBI | | 6-O-Phosphono-b-D-fructofuranose | Generator | | 6-O-Phosphono-β-D-fructofuranose | Generator | | FRUCTOSE-6-phosphoric acid | Generator | | b-D-Fructose 6-phosphate | Generator | | b-D-Fructose 6-phosphoric acid | Generator | | beta-D-Fructose 6-phosphoric acid | Generator | | Β-D-fructose 6-phosphate | Generator | | Β-D-fructose 6-phosphoric acid | Generator |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Weight | 260.1358 |
|---|
| Monoisotopic Molecular Weight | 260.029718526 |
|---|
| IUPAC Name | {[(2R,3S,4S,5R)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | β-D-fructofuranose 6-phosphate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OC[C@@]1(O)O[C@H](COP(O)(O)=O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-2-6(10)5(9)4(8)3(15-6)1-14-16(11,12)13/h3-5,7-10H,1-2H2,(H2,11,12,13)/t3-,4-,5+,6-/m1/s1 |
|---|
| InChI Key | BGWGXPAPYGQALX-ARQDHWQXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose phosphate
- Pentose-5-phosphate
- C-glycosyl compound
- Glycosyl compound
- Monosaccharide phosphate
- Monoalkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Alkyl phosphate
- Tetrahydrofuran
- Secondary alcohol
- Hemiacetal
- Polyol
- Organoheterocyclic compound
- Oxacycle
- Primary alcohol
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-9422000000-bee9b5c8192cd70afd4b | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-9422000000-bee9b5c8192cd70afd4b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0005-9330000000-5c27e76b48f712dafdf1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0540-8097650000-bd7cf45abb555a5c08da | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0002-9200000000-890d2bc5c099d333bb87 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0a4i-0090000000-9e5a9a987b6541abb2c7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0ab9-0000509000-ba24f5137a9692d147ab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-2980000000-6b021fc643037857d79c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dj-6950000000-9aeea6f58646b1ddfd2d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00ds-9300000000-163fede30ff57a757356 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6r-7390000000-3b9471399e651e34edcb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-107d49541ec3b4b1213f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-9182363b16cd6b3e720b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-3090000000-7ae40fd439a28bd2393e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9010000000-53fac253975be4bb66f7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dj-1790000000-cf4773c8e21b0c55570e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9810000000-21980e2a6a612b295b9d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000t-9000000000-a08dc200cb22f420d5bc | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|