| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:07:34 UTC |
|---|
| Update Date | 2020-05-21 16:28:47 UTC |
|---|
| BMDB ID | BMDB0004063 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
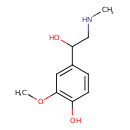
| Common Name | Metanephrine |
|---|
| Description | Metanephrine, also known as metanephrine, belongs to the class of organic compounds known as methoxyphenols. Methoxyphenols are compounds containing a methoxy group attached to the benzene ring of a phenol moiety. Metanephrine is possibly soluble (in water) and a very strong basic compound (based on its pKa). Metanephrine participates in a number of enzymatic reactions, within cattle. In particular, Metanephrine can be converted into 3-methoxy-4-hydroxyphenylglycolaldehyde; which is mediated by the enzyme amine oxidase [flavin-containing] a. In addition, Metanephrine and pyrocatechol can be biosynthesized from epinephrine and guaiacol; which is mediated by the enzyme catechol O-methyltransferase. In cattle, metanephrine is involved in the metabolic pathway called the tyrosine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (+/-)-metanephrine | HMDB | | 3-Methoxy-adrenaline | HMDB | | 3-Methoxyadrenaline | HMDB | | 3-O-Methyl-adrenaline | HMDB | | 3-O-Methylepinephrine | HMDB | | 4-Hydroxy-3-methoxy-N-methylphenethanolamine | HMDB | | DL-3-O-Methyladrenaline | HMDB | | DL-Metanephrine | HMDB | | m-O-Methyladrenaline | HMDB | | Metadrenaline | HMDB |
|
|---|
| Chemical Formula | C10H15NO3 |
|---|
| Average Molecular Weight | 197.231 |
|---|
| Monoisotopic Molecular Weight | 197.105193351 |
|---|
| IUPAC Name | 4-[1-hydroxy-2-(methylamino)ethyl]-2-methoxyphenol |
|---|
| Traditional Name | (+/-)-metanephrine |
|---|
| CAS Registry Number | 5001-33-2 |
|---|
| SMILES | CNCC(O)C1=CC(OC)=C(O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C10H15NO3/c1-11-6-9(13)7-3-4-8(12)10(5-7)14-2/h3-5,9,11-13H,6H2,1-2H3 |
|---|
| InChI Key | JWJCTZKFYGDABJ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as methoxyphenols. Methoxyphenols are compounds containing a methoxy group attached to the benzene ring of a phenol moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Phenols |
|---|
| Sub Class | Methoxyphenols |
|---|
| Direct Parent | Methoxyphenols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Methoxyphenol
- Phenoxy compound
- Anisole
- Phenol ether
- Methoxybenzene
- Alkyl aryl ether
- 1-hydroxy-2-unsubstituted benzenoid
- Aralkylamine
- Monocyclic benzene moiety
- Secondary alcohol
- 1,2-aminoalcohol
- Secondary amine
- Ether
- Secondary aliphatic amine
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Alcohol
- Aromatic alcohol
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-014i-0910000000-a886bef599ae583766b9 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-0910000000-a886bef599ae583766b9 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9300000000-99e319cf43defe7cc611 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0g6r-9553000000-f11ff40fb7a8af16bb25 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-0900000000-4c31604c1b2a54419f92 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001j-0900000000-12556e41dd73a4575162 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6w-4900000000-403de00b0b43ed22918f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-3d29615883725ba8e07c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0032-1900000000-1ccaa0d8ce9764cdfe35 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pic-4900000000-051892c7895245a3de0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-0900000000-545ae65ccf6b1aaf8f43 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001a-1900000000-ad33b1d4593df03f60e1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-5900000000-61cd5865cf7abd2bba67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-33b506e73b8f2abe6bdb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000b-0900000000-361e5b74103dd493ac47 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4m-6900000000-49bdcf483f24e0b4ffd4 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|